Get binding affinities from almost nothing

MicroScale Thermophoresis (MST) is the only way to get a direct binding affinity in solution, with high accuracy, and – most importantly – while using practically no sample at all (down pg of protein per experiment).
2bind’s MST assays can be scaled from just a single interaction up to 10.000’s of interactions.
MST works with almost all classes of target molecules as long as they can be made fluorescent. The variety of ligand molecules is even greater – from molecular fragments up to virus-like particles. The fluorescent target molecule is kept at a constant concentration and the non-fluorescent ligand molecule is titrated across a large concentration range.
Affinity
LIGAND BINDING
KD
Equilibrium dissociation constant. Can be obtained by kinetic or classical equilibrium binding analysis. Provides information about the strength but not the dynamics of an interaction.
ΔH
Binding enthalpy. KD values at different temperatures can be used to obtain the binding enthalpy of an interaction via vant-Hoff-plots.
Ligand scouting
Fast screening for yes/no binding answers.
Other assay types
COMPETITION ASSAYS
Determination of competition of several ligands for a selected binding site on a target.
COMPLEX INHIBITION ASSAYS
Determination of binding site blocking by a ligand and prevention of binding of a natural interaction partner.
STOICHIOMETRY ASSAYS
Determination of equilibrium binding stoichiometry of target-ligand complex formation.
What MST can tell
Single-dose screen
MST will tell you whether a target and ligand generally interact from a single-dose experiment.
Steady-State Affinity
Protein Aggregation
MST will tell you whether your protein samples aggregate or if aggregation is induced by ligand binding.
Ligand competition
MST will tell you competitive effects between multiple binders of your targets.
- Drug discovery (compound-based and fragment-based)
- Drug discovery (protein-targeted and RNA-targeted)
- Protein characterization
- Additive screening
- Enzyme-inhibitor screening
- Drug discovery (compound-based and fragment-based)
- Drug discovery (protein-targeted and RNA-targeted)
- Protein characterization
- Additive screening
- Enzyme-inhibitor screening
- Protein and enzyme quality control
- Buffer screening and optimization
- Protein storage optimization
- Ligand-induced aggregation testing
- Complex binding analysis
- Mode-of-action analysis
- Competitor screening
- PPI-disruption analysis
Advantages of MST
2bind MST Services
2bind offers a variety of services using the MicroScale Thermophoresis technology. Benefit from fast and precise analysis of molecular interactions.
Enjoy minimal sample consumption (just nM concentrations and µL volumes) and robust analysis methods. The 2bind MST services can be combined as you choose for all questions in drug discovery, antibody development, protein biophysics and analysis, as well as aptamer characterization.
MST Technology and FAQs
Overview
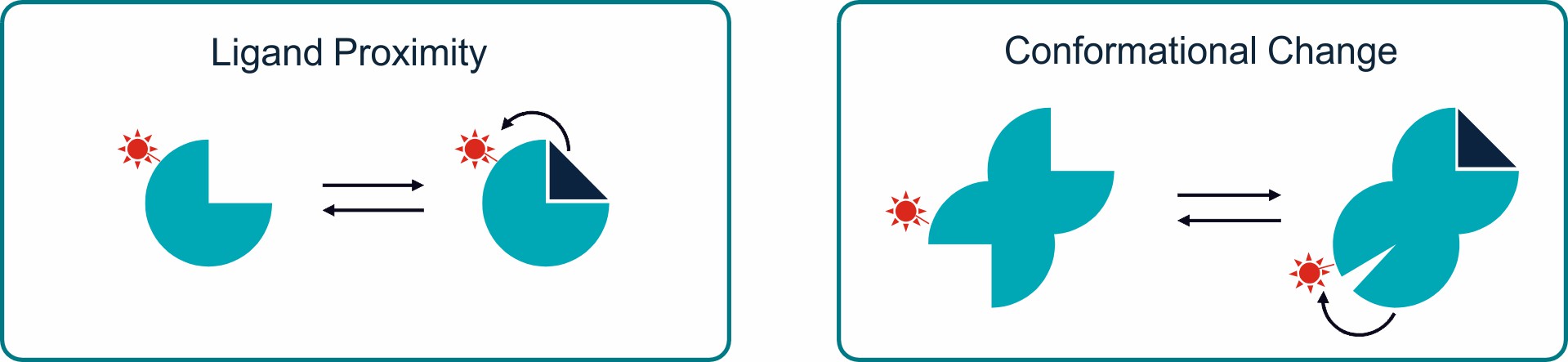
The MST signal is generated by the TRIC effect. TRIC stands for Temperature-Related Intensity-Change and describes how the fluorescence intensity of a fluorophore changes with the local temperature of the solution. For the vast majority of fluorophores, the fluorescence intensity decreases with increasing temperature.
The TRIC effect is strongly related to (even very subtle) changes in the electronic microenvironment of the fluorophore. Such changes mostly happen upon binding of a ligand to the target molecule and/or conformational changes of the target molecule upon binding of a ligand.
Technology
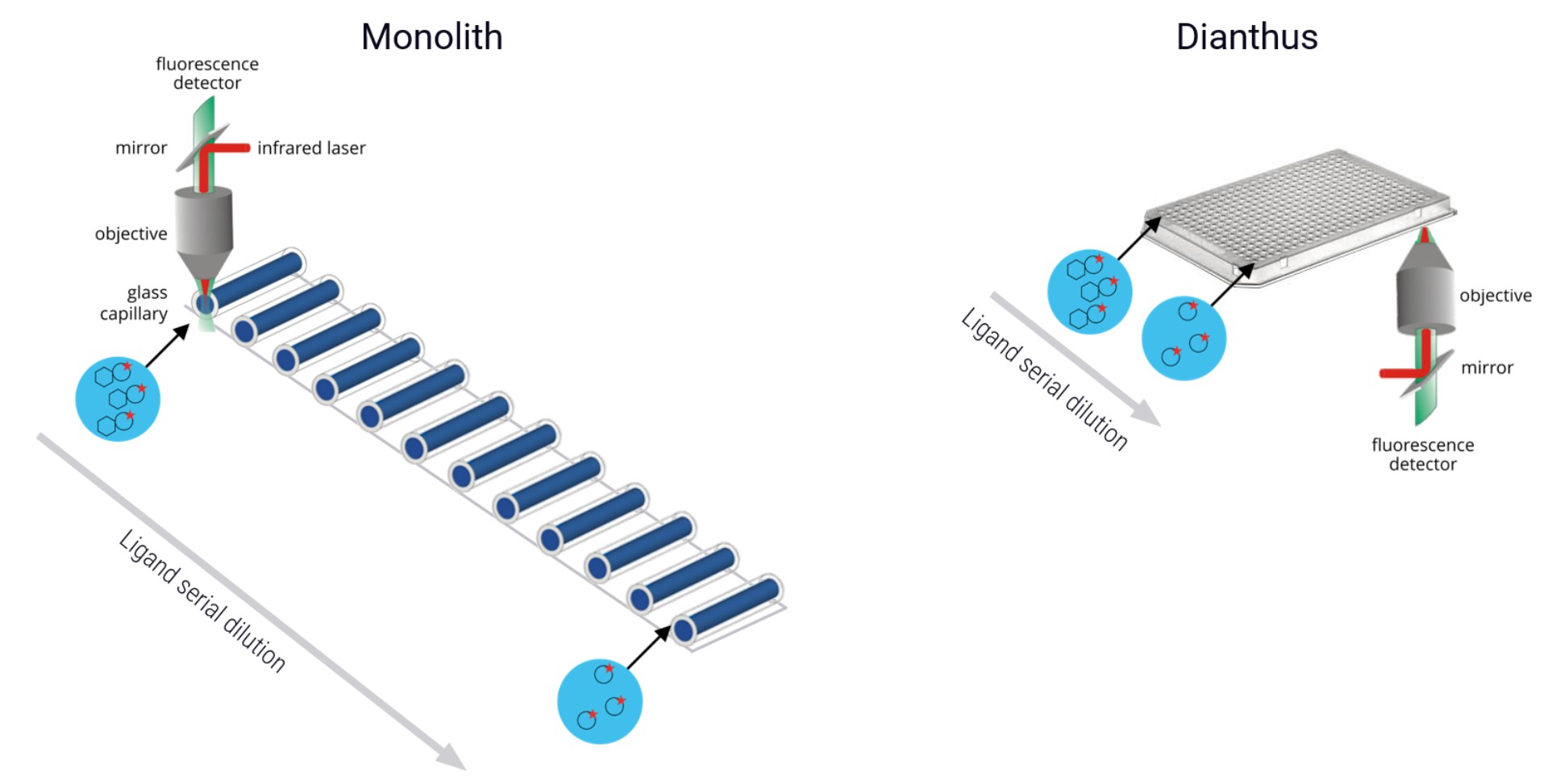
MST measurements are possible either in a glass capillary setup (Monolith-series instruments, NT.Automated) or in a plate-based format (Dianthus). In both setups, an infrared laser is used to generate a precise temperature gradient while an LED is used for the excitation of fluorescent molecules inside the glass capillaries or the plate wells. The capillaries or wells each contain a mix of fluorescent binding partner (usually called the “target” and non-fluorescent binding partner, which is titrated in a dilution series (usually called the “ligand”).
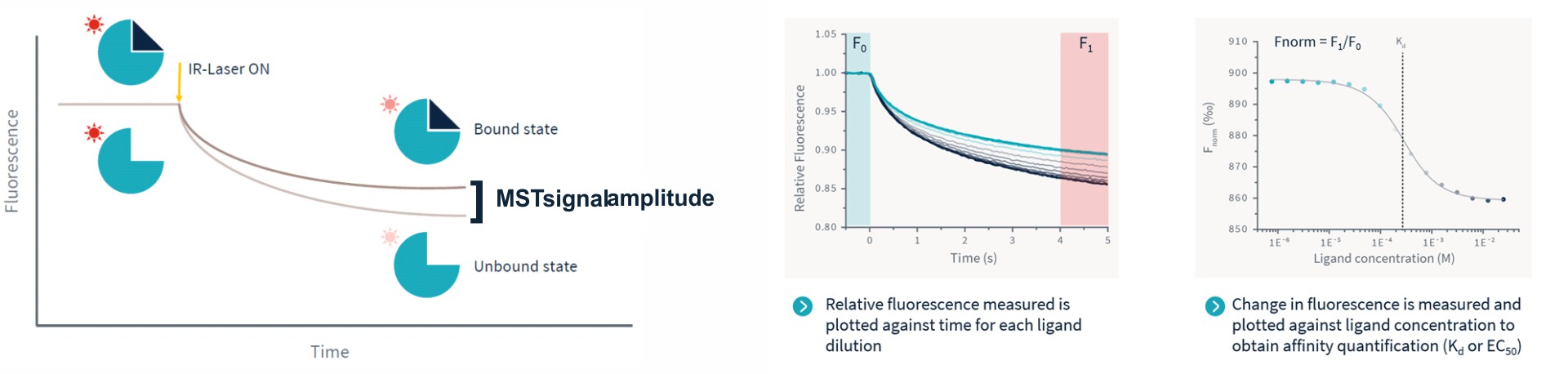
The sample fluorescence in each capillary or well is measured over time. After a certain time, the infrared laser is switched on and the sample is heated. Such resulting MST traces are recorded for each ligand concentration and plotted against time. The calculated Fnorm values from the MST traces (fluorescence in region F1 divided by fluorescence in region F0) are dose-dependent and can be well described by the law of mass action. A plot of Fnorm against ligand concentration then returns the dissociation constant KD of the interaction.
MST allows for the monitoring of either fluorescently-labeled molecules or intrinsically fluorescent molecule (such as proteins; the latter would be a truly label-free measurement). MST measurements are possible in any kind of buffers, even in serum, plasma, cell lysate, urine, mucus, or other environmental matrices. Data generation is fast and precise and the data output is comparable to other biophysical methods.
Typical applications
Typical applications of MicroScale Thermophoresis include:
- Hight-throughput target-ligand interaction screenings
- Steady-state binding affinity assay
- Steady-state binding affinity assay in biological liquids
- Sandwich assays (1 target, 2 ligands)
- Competition assays
- Binding affinity assays with multiple binding partners
For more information, please visit our Application Database. The different service areas can be found under MST Services.
Compatible Fluorescent Dyes
MicroScale Thermophoresis (MST) relies on measuring the fluorescence of the studied molecules. Thus, one of the binding partners has to be fluorescent. Intrinsically fluorescent proteins can be analyzed without the need for further fluorescent labeling.
Depending on which MST Service you require, different fluorescent methods and/or labelings are possible.
Alternatively, molecules with no intrinsic fluorescence can be labeled with fluorophores. Most commonly, the labels listed in the table below are used. 2bind performs the labeling of your target protein for you with all possible dyes. Another option is to fuse a potential target protein with an intrinsically fluorescent protein fluorophore such as GFP, RFP or the like.
In the case of DNA or RNA target molecules direct covalent linkage of fluorophores (e.g. Cy5) has proven well.
The following table gives an overview over the most commonly used fluorophores in MST. Please keep in mind that, in principle, every fluorophore can be used as long as its excitation and emission wavelength ranges match the ones of the fluorophores listed here.
| Fluorophore | Excitation (nm) | Emission (nm) |
|---|---|---|
| BCECF | 480 | 525 |
| GFP | 488 | 507 |
| NT-495 (BLUE) | 493 | 521 |
| Fluorescein (FITC) | 495 | 519 |
| Alexa488 | 495 | 519 |
| YFP | 514 | 527 |
| Alexa532 | 530 | 555 |
| T AMRA | 546 | 579 |
| Cy3 | 550 | 570 |
| RFP | 555 | 584 |
| NT-547 (GREEN) | 557 | 574 |
| Alexa546 | 560 | 672 |
| Cy5 | 649 | 670 |
| NT-647 (RED) | 650 | 680 |
| Alexa647 | 652 | 668 |
| NT-RED 2nd generation | 650 | 670 |
Label-free MST
Due to tryptophan residues in their amino acid sequence, proteins can be intrinsically fluorescent. The intrinsic fluorescence of such proteins can be used to monitor their thermophoretic movement. By that no interaction partner has to be modified and the MicroScale Thermophoresis (MST) assay is truly label-free.
Such label-free MST is often interesting for specific protein MST services, whose intrinsic fluorescence can be used as an alternatively to a fluorescent label.
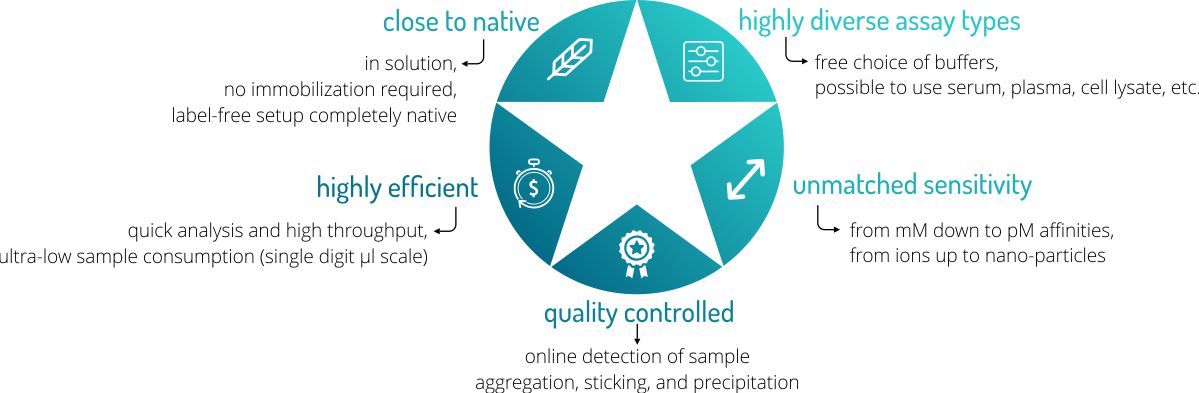
Advantages
MicroScale Thermophoresis (MST) offers a number of great advantages over other biophysical methods for determining the affinity of a molecular interaction. Follow this link for an overview over our possible services.
- Low sample consumption → minimum of only 6 µl is required per sample
- Free choice of assay buffers → also biological liquids possible such as serum or cell lysate
- Very short analysis time → short analysis time enables high throughput
- Real-time quality controls → online aggregation, precipitation, and sticking controls
- Wide temperature range → analysis possible from 20°C to 45°C
- No immobilization required → measurement is done truly in solution
- Wide concentration range → affinities can be analyzed in the pM-mM range
- Wide molecule size range → from 100 Da to 1 MDa
For a comparison of MST with other biophysical techniques, please refer to our Technology Comparison Guide.
FAQ – General
What kinds of molecular interactions can I measure?
You can measure bi-molecular interactions between any kind of molecule: protein, RNA, DNA, small molecule compounds, lipids, carbohydrates (See figure on top of this page).
Is it possible to analyze interactions with aptamers?
More information on this topic is available in our Aptamer Services.
What information do I get from an MST measurement?
MicroScale Thermophoresis (MST) is not only able to determine the affinity and binding strength of a molecular interaction, but also allows for assessing other physical parameters such as stoichiometry, aggregation, precipitation, enthalpy (van’t Hoff plot), slow enzyme kinetics, and oligomerization.
For specialized kinetics measurements, consider Biolayer Interferometry. If you are interested in the thermodynamics of a molecular interaction, Isothermal Titration Calorimetry is an option. For an in-depth analysis of protein aggregation, protein stability and protein unfolding, take a look at the nanoDSF technique.
What type of fluorescent dyes can I use?
Is it possible to measure without labelling a molecule?
Can I measure binding kinetics with MicroScale Thermophoresis?
An in-depth kinetics analysis of molecular interactions is possible with Biolayer Interferometry.
Can I measure thermodynamics with MicroScale Thermophoresis?
An in-depth thermodynamics analysis of a molecular interaction is possible with Isothermal Titration Calorimetry.
FAQ – Samples
Can I check for the quality of my samples?
An in-depth analysis of the stability and aggregation behavior of your protein samples can be done with nanoDSF.
Is it possible to measure the binding of small molecules?
What are the required concentrations for small molecules?
My protein is only stable at high ionic strength. Can I still test for binding with MST?
If you are not sure whether your sample protein really requires high ionic strength buffers, it is possible to investigate protein stability, thermal unfolding, and its aggregation behavior using nanoDSF. We offer a variety of Services in this respect.
I want to measure binding to a Mega-Dalton (MDa) molecule. Is this possible?
FAQ – Assay Conditions
Are there any limitations to the buffers or additives I can use?
A great way to check the tolerance of your protein samples for a vast number of additives is nanoDSF, a method that analyzes the thermal stability and unfolding of proteins. This technique can be used for buffer and additive screenings.
Can I measure in biological liquids like serum or lysates?
If you are interested in the stabilty of your protein sample, this can be determined using our nanoDSF technology.