Binding kinetics of antibodies to the HIV Env glycoprotein measured with Biolayer Interferometry (BLI)
Benjamin Zimmer, Corinna Popp, Maximilian Plach
2bind GmbH, Regensburg, Germany
BLI Technology
Biolayer Interferometry (BLI) measures binding kinetics. To do this, white light is sent down an optical glass fiber and is reflected from two optical layers (Figure 1, left): First, an internal reference layer, where reflection occurs at the interface between the glass fiber and the optical layer. Second, a bio-compatible layer, where reflection occurs at the interface with the surrounding liquid. Reflected light interferes either constructively or destructively at different wavelengths, resulting in a distinctive interference. While the light from the reference layer remains constant throughout an experiment, the attachment or detachment of molecules to/from the sensor surface changes the thickness of the bio-compatible layer and thereby the path length of the reflected light, resulting a spectral shift in the interference pattern (Figure 1, right) (Concepcion et al., 2009; Fortebio Application Note 14).
Therefore, BLI allows for observing target-ligand association and dissociation in real-time. Information from the binding curves can be used to determine binding parameters such as the association and dissociation rates constants (ka, kd) or the equilibrium dissociation constant (KD).
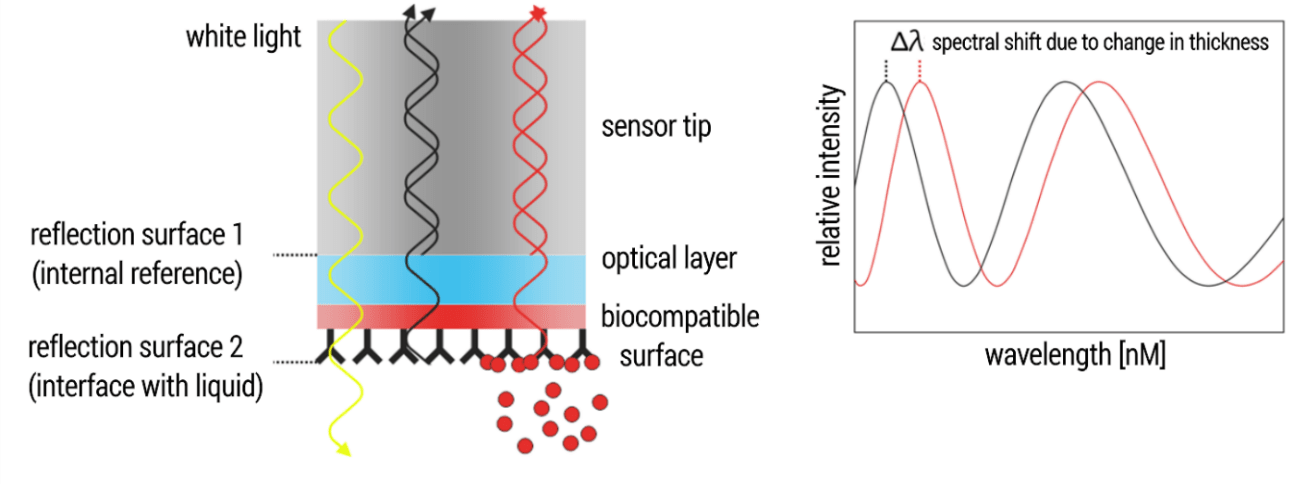
Figure 1. Technical BLI setup.
Introduction: Broadly-neutralizing anti-HIV antibodies and their binding kinetics
Broadly neutralizing antibodies (bnAbs) play a central role in the development of preventive and therapeutic vaccines against the human immunodeficiency virus 1 (HIV-1; Sok & Burton, 2018). They efficiently neutralize the fusion machinery of HIV-1, which the virus needs for entering human cells. One integral component of this fusion machinery is the trimeric glycoprotein Env. For development of bnAB-based HIV-1 vaccines, detailed information about the interactions between Env and the bnAbs, ideally including affinity and kinetic parameters, is paramount. In order to demonstrate the performance of BLI for such questions, we studied the interaction between a soluble, stabilized Env (BG505 SOSIP.664) and two Env-directed bnAbs (2G12, PGT145; Sanders et al., 2013; Murin et al., 2014; Lee et al., 2017).
Results 1: Binding kinetics of BG505 SOSIP.664 and bnAb 2G12
The bnAb 2G12 has the unique feature that its two Fab arms assemble into one rigid Env-binding unit. Thus, both paratopes together engage one Env epitope. In total, three 2G12 antibodies bind to one Env trimer. With the aim to analyze the interaction between 2G12 and BG505 SOSIP.664 trimers under a 1:1 binding situation, the trimeric Env variant BG505 SOSIP.664 was used as the ligand (immobilized to the sensor surface) and the bnAb 2G12 was titrated as the analyte. We compared two immobilization strategies: First, immobilization of biotinylated BG505 SOSIP.664 to Streptavidin sensors. Second, capturing of His-tagged BG505 SOSIP.664 trimers to Ni-NTA sensors. Both immobilization strategies are pictured in Figure 2.

Figure 2. Experimental setup for kinetic characterization of the interaction between BG505 SOSIP.664 and bNAb 2G12.
1) Sensor hydration, 2) Immobilization of biotinylated BG505 SOSIP.664 to Streptavidin sensors or alternatively, capturing of His-tagged BG505 SOSIP.664 trimers to Ni-NTA sensors, 3) 2G12 association, and 4) 2G12 dissociation.
The observed sensorgrams (Figure 3) showed single-exponential association phases, with some heterogeneity only at the highest analyzed analyte concentration. This, together with the good fit quality (R2 > 0.95, low X2), indicated that the interaction was analyzed under the desired 1:1 binding situation. In addition, both immobilization strategies delivered comparable results regarding ka, kd and KD.
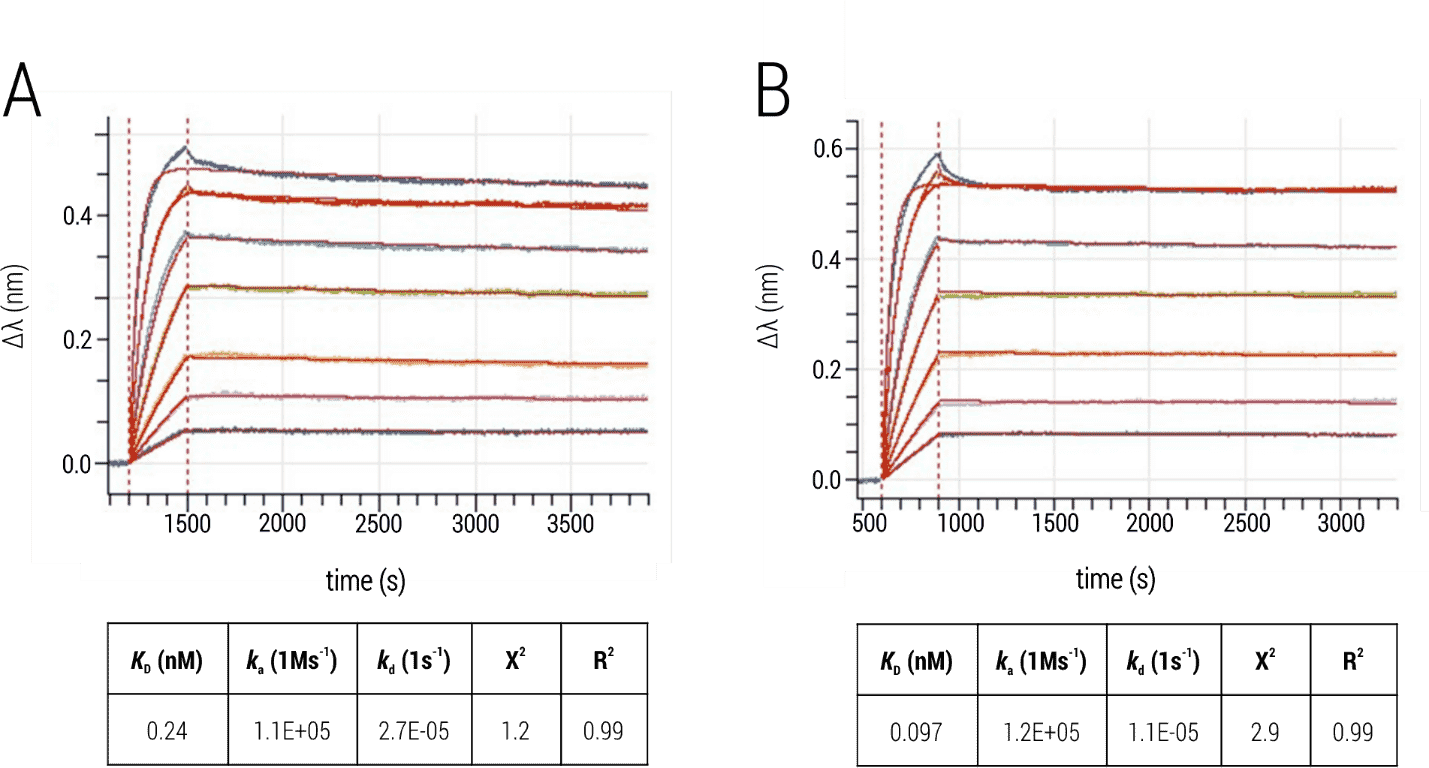
Figure 3: Interaction of BG505 SOSIP.664 trimers and bnAb 2G12. (A) Immobilization of the Env trimer to Ni-NTA sensors. (B) Immobilization of biotinylated Env trimer to Streptavidin sensors. The 2G12 was titrated and kinetic parameters were obtained by fitting processed data with the 1:1 binding kinetics model (global fit; red).
Results 2: Binding kinetics of BG505 SOSIP.664 and bnAb PGT145
The bnAb PGT145 engages a quaternary epitope that is present only once per Env trimer. Thus, in order to analyze the interaction under a 1:1 binding situation, the PGT145 bnAb was immobilized as the ligand and BG505 SOSIP.664 trimers were titrated as the analyte. Two different immobilization strategies were tested: First, immobilization of biotinylation PGT145 on Streptavidin sensors. Second, capturing of PGT145 on anti-human IgG-Fc capture sensors (AHC sensors). Both immobilization strategies are pictured in Figure 4.
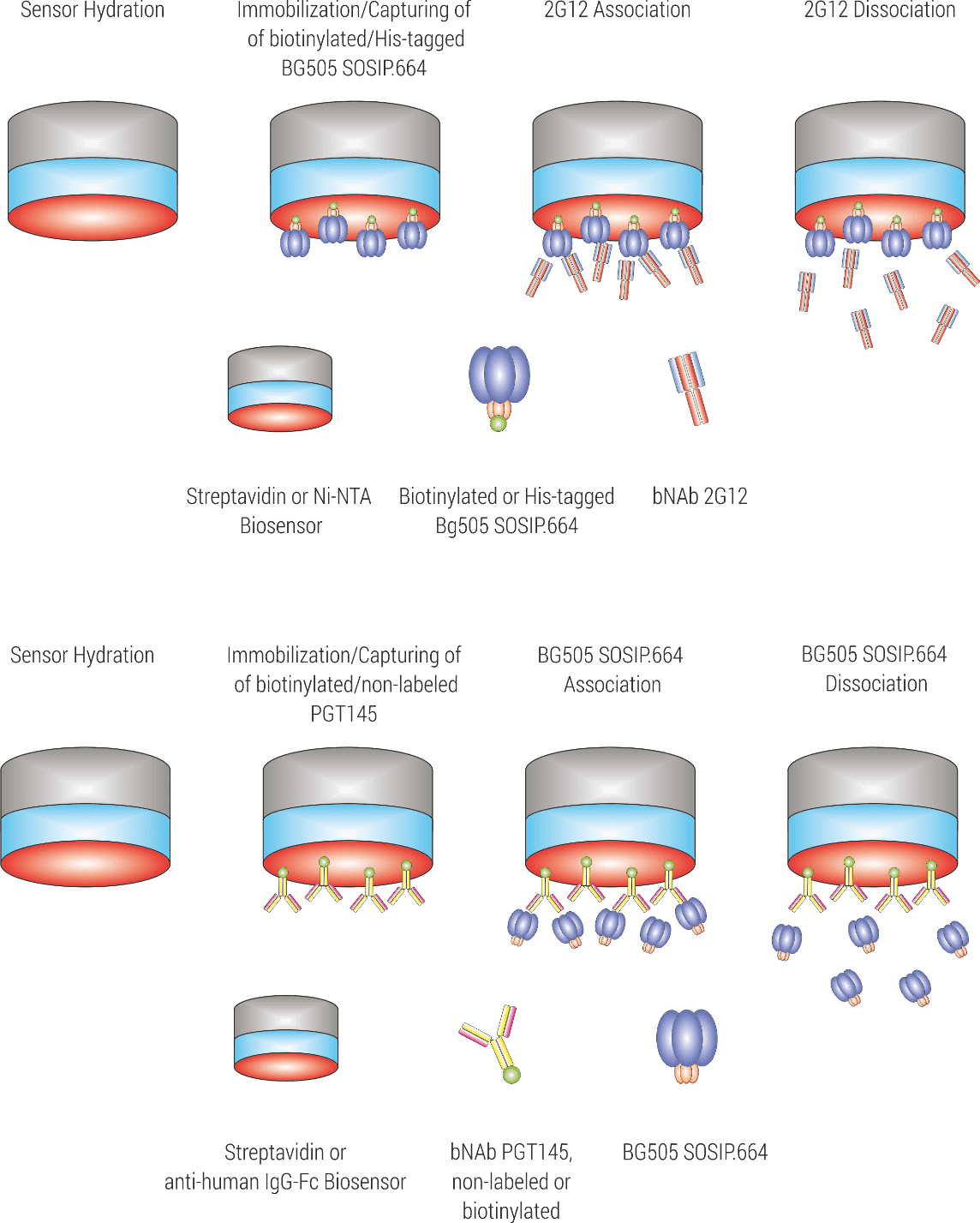
Figure 4. Experimental setup for kinetic characterization of the interaction between BG505 SOSIP.664 and bNAb PGT145.
1) Sensor hydration, 2) Immobilization of biotinylated PGT145 to Streptavidin sensors, or alternatively, capturing of PGT145 to anti-human FC sensors, 3) BG505 SOSIP.664 association, and 4) BG505 SOSIP.664 dissociation.
The observed sensorgrams (Figure 5) showed the single-exponential association phase of a classical 1:1 interaction, supported by good fit quality. Calculated ka, kd and KD-values were comparable, validating the feasibility of the two different immobilization strategies.
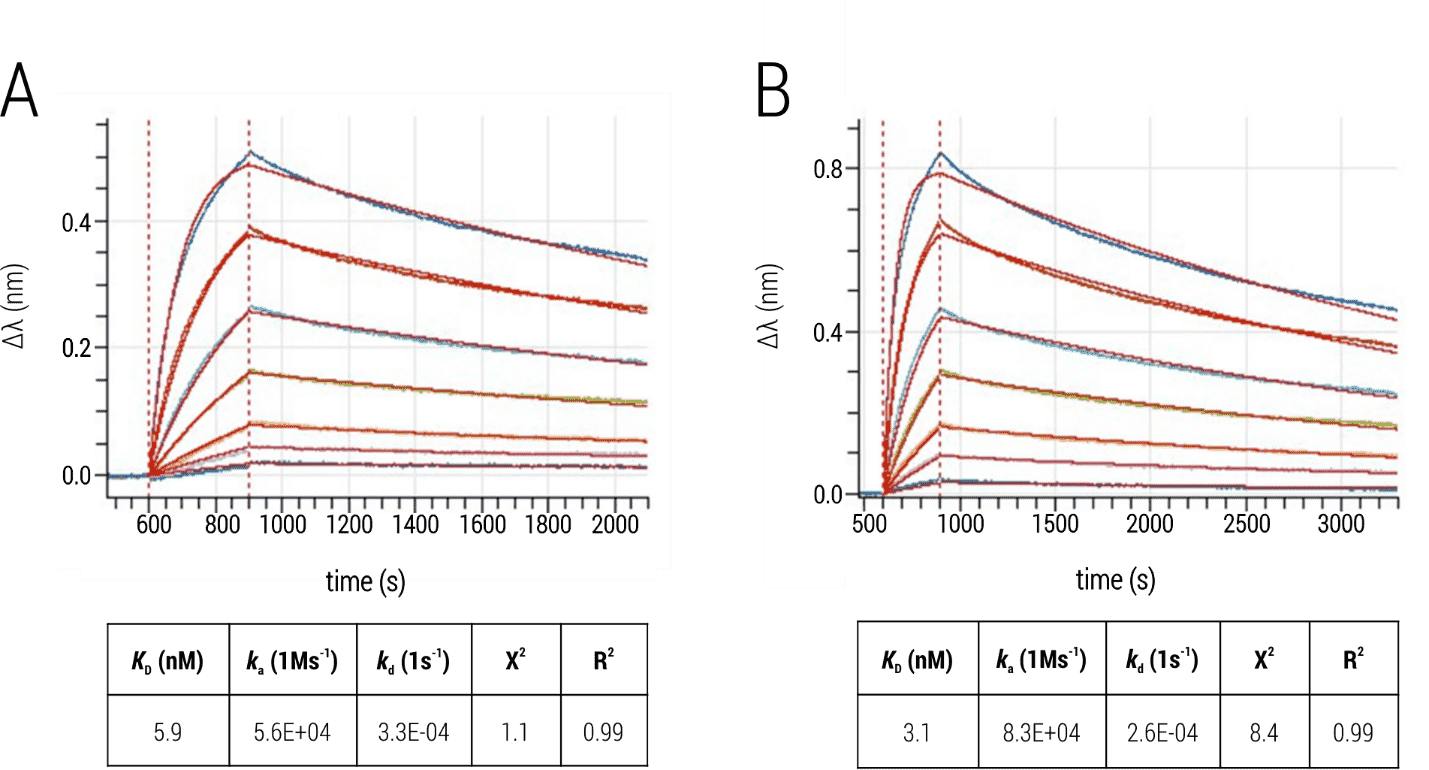
Figure 5: Interaction of BG505 SOSIP.664 trimers and bnAb PGT145. (A) Immobilization of the biotinylated bnAb to Streptavidin sensors. (B) Immobilization of PGT145 to AHC sensors. The Env trimer was titrated and kinetic parameters were obtained by fitting processed data with the 1:1 binding kinetics model (global fit; red).
Conclusion: 2bind BLI assay for measuring binding kinetics of antibody-HIV interactions
Considering the significantly different binding characteristics of bNAbs 2G12 and PGT145 to the HIV-1 protein Env, we successfully established BLI setups that allowed the kinetic characterization of these antibodies to their trimeric antigen under a 1:1 binding situation. In addition, we could show that different immobilization strategies delivered comparable results for the respective antibody-Env interaction regarding kinetic parameters and binding affinities.
Click here for a full list of services for antibody analysis. If you like to directly contact us for any kind of service, use the contact form below.
Literature
Concepcion et al. Label-free detection of bio-molecular interactions using BioLayer interfero-metry for kinetic characterization. Comb. Chem. High Throughput Screen. 2009, 12(8): 791–800.
Fortébio. Application Note 14: Biomolecular Binding Kinetics Assays on the Octet Platform. Available at: https://www.fortebio.com/. (Accessed: 8th March 2019)
Lee et al. A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic β-Hairpin Structure. Immunity. 2017, 46(4): 690-702
Murin et al. Structure of 2G12 Fab2 in Complex with Soluble and Fully Glycosylated HIV-1 Env by Negative-Stain Single-Particle Electron Microscopy. J. Virol. 2014, 88(17): 10177-10188.
Sanders et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutra-lizing but not non-neutralizing antibodies. Plos Pathog. 2013, 9(9):e1003618.
Sok & Burton. Recent progress in broadly neutra-lizing antibodies to HIV. Nat. Immunol. 2018, 19(11):1179-1188.
