Viral RNA as targets for drug discovery: Analyzing HIV Tat-Tar inhibitors with Microscale Thermophoresis (MST)
Ákos Harkai, Thomas Schubert
2bind GmbH, Regensburg, Germany
MST Technology
MicroScale Thermophoresis (MST) is an optical fluorescent method. It records the changes in fluorescence of a target molecule as a function of temperature and the concentration of the cognate ligand molecule. Thus, in MST, the target must be fluorescent and the ligand must not be fluorescent in the same wavelength range as the target. The fluorescence changes are the result of two effects:
First, molecules exhibit directed movement along temperature gradients in solution, an effect called thermophoresis (Duhr & Braun, 2006). Such thermophoretic movement of a molecule is characterized by its size (hydrodynamic radius), its surface charge, and its hydration shell. All three parameters can be affected by binding of a ligand molecule. Thus, a concentration-dependent titration of the target molecule’s ligand induces changes in the thermophoretic movement. These changes translate in quantifiable, spatial fluorescence changes, which are easily tracked optically via the target fluorescence.
Second, fluorescence is a function of the temperature. Thus, the fluorescence of the target molecule varies with the temperature (Baaske et al., 2010). This temperature-dependence is additionally affected by the local molecular surroundings of the fluorophore. Thus, binding of a ligand molecule to the fluorescent target can change the chemical environment of the target fluorophore and thus the overall detected temperature-dependence of the fluorescence. This effect is known as TRIC (temperature-related intensity change; Gupta, Duhr & Baaske, 2018).
The thermophoresis and TRIC signals are additive and thus both contribute to the high sensitivity and robustness of MST measurements towards molecular binding events of all kinds. Thus, MST can be used for determining the affinity and binding strength of almost any kind of molecular interaction with very low sample consumption and very high sensitivity.
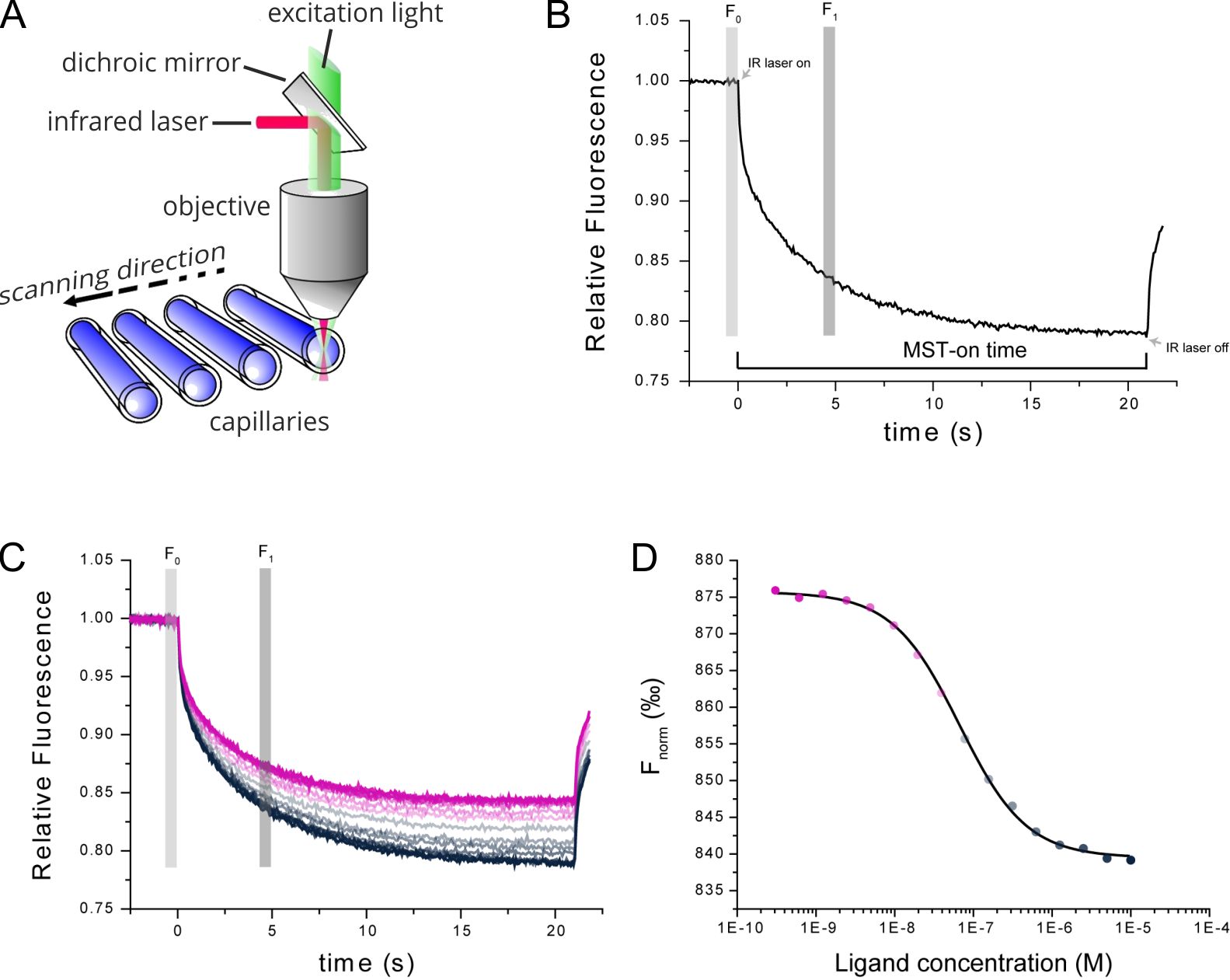
Figure 1. Technical MST setup. (A) MST measurements take place in small glass capillaries. Infrared and fluorescence lasers are used for generation of the MST effect and sample tracking. (B) TRIC and thermophoresis together account for a time-dependent change in fluorescence upon infrared-heating of the sample capillaries. (C) Multiple MST traces are recorded for different mixture ratios of target and ligand molecules. (D) Dose-response analysis of the MST traces allows for determination of the steady-state affinity of the target-ligand interaction.
Introduction: RNA as targets for drug discovery
The appearance of new viral diseases as well as the still limited number of available medications pose a need for new antiviral agents (Graci & Cameron, 2008). Extending the search for such new antivirals from classical protein targets to other molecular structures of viruses, like important RNA or DNA strands, demands for biophysical characterization methods that not only work excellently with small chemical compounds but also with a variety of biological macromolecule classes. Here, we demonstrate the power of custom-made MicroScale Thermophoresis (MST) assays for testing the potency of new small molecule drug candidates to a viral RNA target.
The Trans-activation response (TAR) element is a well-characterized region in the HIV-1 genome. The element forms a stem-loop secondary structure, which serves as specific binding site for the HIV Trans-activator of transcription (Tat) protein. The purpose of the Tat-TAR complex is to speed up the viral reproduction process in infected host cells, taking advantage of their expression apparatus (Debaisieux et al., 2012). Interrupting Tat-TAR complex is one therapeutic strategy in the treatment of HIV infection. Mitoxantrone and some aminoglycosides are known potent agents, which can inhibit the formation of Tat-TAR complex via binding to various regions of TAR RNA. (Stelzer et al., 2011) Here, an instructive MST comparison was performed in order to find out the binding affinity of four different inhibitor candidates.
The MST assay was performed with the TAR RNA as the target molecule, meaning it was synthesized with a site-specific label. By that it could directly be used for MST. The different inhibitor candidates were titrated across a large concentration range in order to determine their steady-state affinity to TAR. In order to validate the assay, the native binder Tat was used as a control.
Results: Selectivity of small molecule inhibitors of HIV Tar RNA
The Tat-TAR complex showed an extremely high affinity with a KD-value in the pM concentration range (KD= 0.23 ± 0.17 nM). Moreover, mitoxantrone also displayed high affinity towards TAR RNA with a KD-value of 56.3 ± 12.3 nM. In comparison, neomycin and netilmicin demonstrated a lower affinity with a KD-values of 3.5 ± 0.7 µM and 294 ± 48 µM. Finally, Paneticin did not interact with TAR RNA despite being a neomycin analogue (Figure 2).
Importantly, the full inhibitor panel characterization was performed with only minimum amounts of the expensive, fluorescently labeled TAR RNA (single-digit nM concentration, just 100 µL per assay) and also low volumes of the chemical molecules (single-digit µL of 10 mM DMSO stocks).
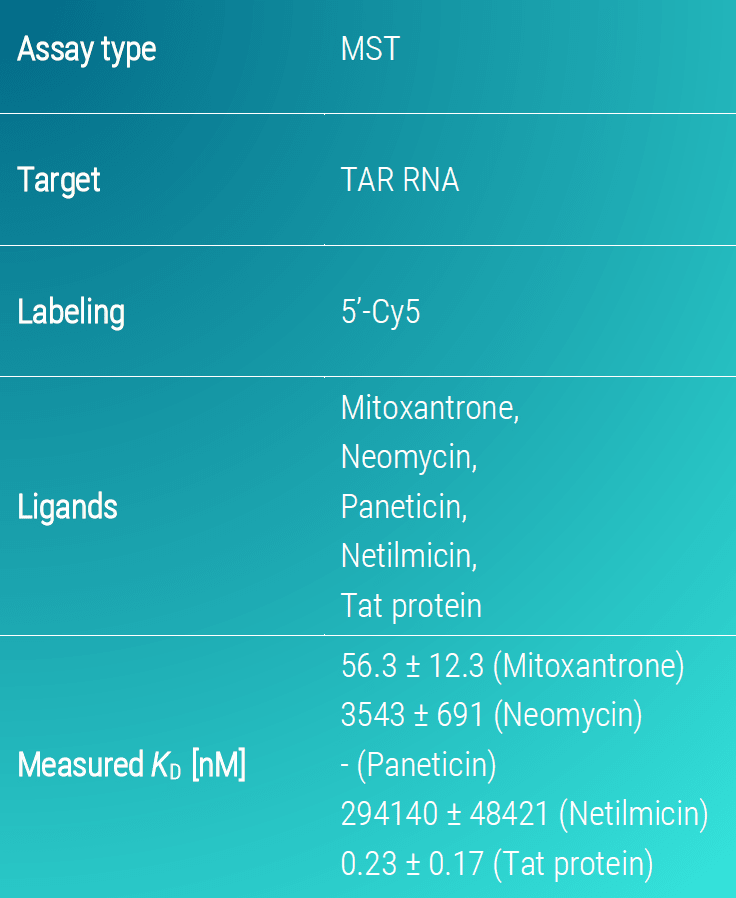
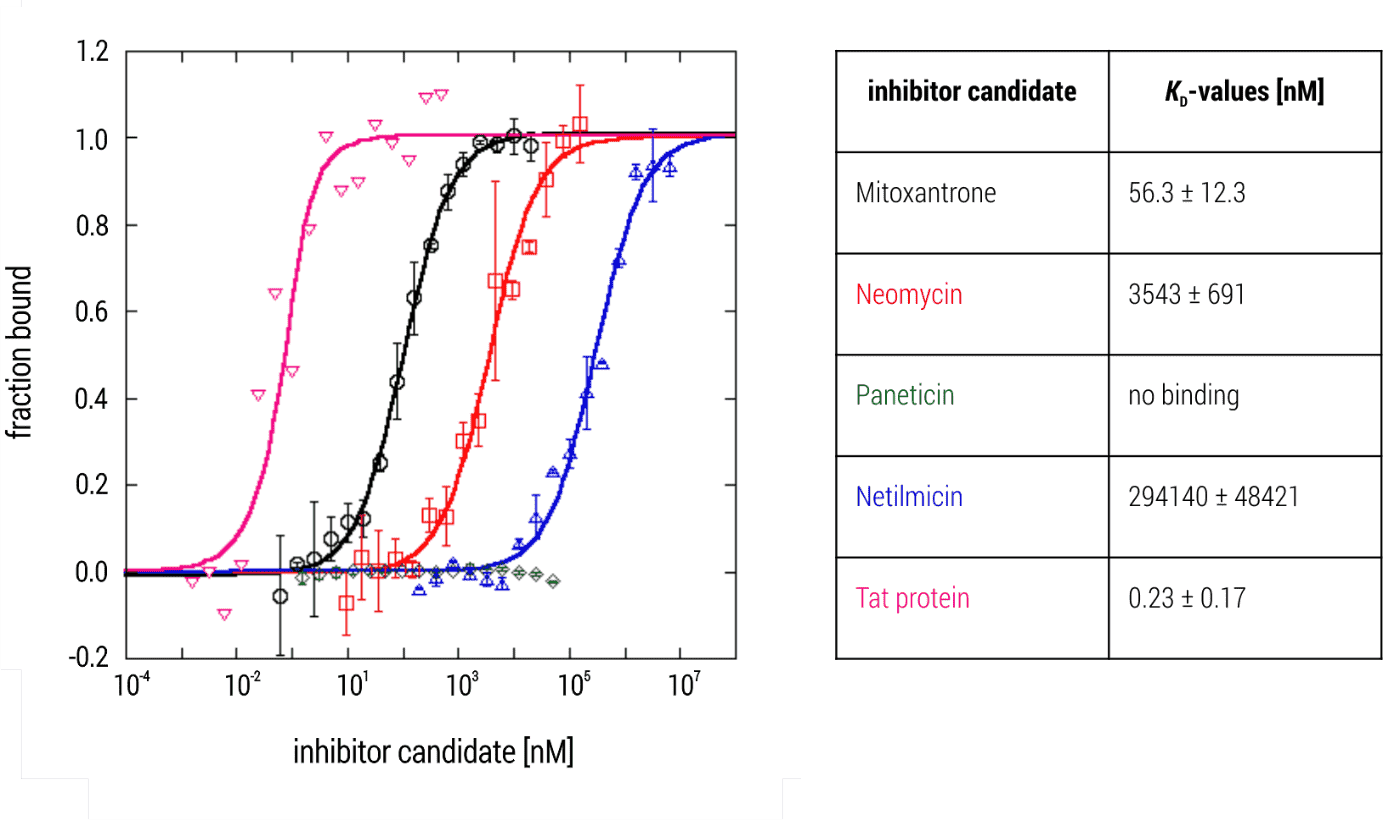
Figure 2. MST binding isotherms of TAR RNA and various small molecule inhibitor candidates. Data are the mean and standard deviation of two independent experiments and are fitted to a KD‑binding model under a 1:1 binding situation. Tat protein served as a control for establishing the experiment.
Conclusion: 2bind MST assays for small molecule – RNA interactions
In this application note, we demonstrated that our versatile MST assays are capable of measuring interactions between a sensitive viral RNA and its small molecule ligand compounds as well as its natural binding partner. With only minor tweaks, such an RNA-interaction assay can be adapted for almost all RNA-small molecule interactions.
Click here for a full list of MST services. If you like to directly contact us for any kind of service, use the contact form below.
Literature
Baaske et al. Optical thermophoresis for quantifying the buffer dependence of aptamer binding. Angew. Chem. Int. Ed Engl. 2010; 49(15): 2238-2241
Debaisieux et al. The Ins and Outs of HIV‐1 Tat. Traffic. 2012; 13 (3): 355–63.
Duhr & Braun. Why molecules move along a temperature gradient. Porc. Natl. Acad. Sci. USA 2006; 103(52): 19678-19682.
Graci & Cameron. Therapeutically targeting RNA viruses via lethal mutagenesis. Future Virol. 2008; 3(6): 553–566.
Gupta, Duhr & Baaske. MicroScale Thermophoresis (MST). Encyclopedia of Biophysics. 2018; 1–5
Stelzer et al. Discovery of Selective Bioactive Small Molecules by Targeting an RNA Dynamic Ensemble. Nat Chem Biol. 26; 7(8): 553–559.
