Label-free and in-solution measurement of protein-ion interactions
Clemens Entzian, Thomas Schubert
2bind GmbH, Regensburg, Germany
MST Technology
MicroScale Thermophoresis (MST) is an optical fluorescent method. It records the changes in fluorescence of a target molecule as a function of temperature and the concentration of the cognate ligand molecule. Thus, in MST, the target must be fluorescent and the ligand must not be fluorescent in the same wavelength range as the target. The fluorescence changes are the result of two effects:
First, molecules exhibit directed movement along temperature gradients in solution, an effect called thermophoresis (Duhr & Braun, 2006). Such thermophoretic movement of a molecule is characterized by its size (hydrodynamic radius), its surface charge, and its hydration shell. All three parameters can be affected by binding of a ligand molecule. Thus, a concentration-dependent titration of the target molecule’s ligand induces changes in the thermophoretic movement. These changes translate in quantifiable, spatial fluorescence changes, which are easily tracked optically via the target fluorescence.
Second, fluorescence is a function of the temperature. Thus, the fluorescence of the target molecule varies with the temperature (Baaske et al., 2010). This temperature-dependence is additionally affected by the local molecular surroundings of the fluorophore. Thus, binding of a ligand molecule to the fluorescent target can change the chemical environment of the target fluorophore and thus the overall detected temperature-dependence of the fluorescence. This effect is known as TRIC (temperature-related intensity change; Gupta, Duhr & Baaske, 2018).
The thermophoresis and TRIC signals are additive and thus both contribute to the high sensitivity and robustness of MST measurements towards molecular binding events of all kinds. Thus, MST can be used for determining the affinity and binding strength of almost any kind of molecular interaction with very low sample consumption and very high sensitivity.
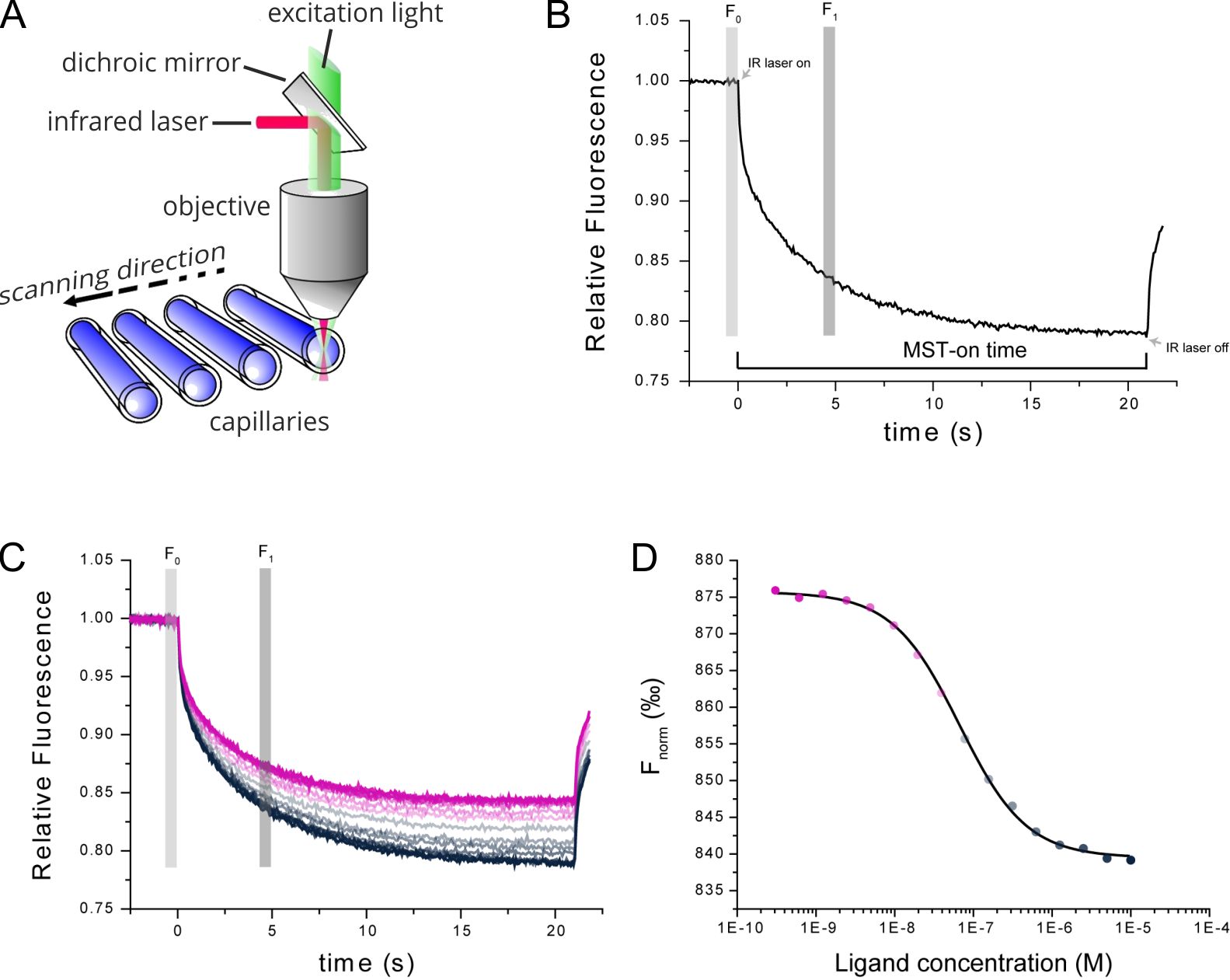
Figure 1. Technical MST setup. (A) MST measurements take place in small glass capillaries. Infrared and fluorescence lasers are used for generation of the MST effect and sample tracking. (B) TRIC and thermophoresis together account for a time-dependent change in fluorescence upon infrared-heating of the sample capillaries. (C) Multiple MST traces are recorded for different mixture ratios of target and ligand molecules. (D) Dose-response analysis of the MST traces allows for determination of the steady-state affinity of the target-ligand interaction.
Introduction: Interactions between proteins and small ions
A great number of enzymatic reactions are dependent on cofactor-binding. However, an analysis of the interaction between enzymes and their cofactors in detail is challenging. Often, it is complicated to access the basic binding parameters such as the affinity of ions using the state-of-the-art biophysical methods. Here, we demonstrate how MST can be used to test the ion-dependence of a target enzyme.
Micrococcal nuclease (MNase) is a Ca2+-dependent endo-exonuclease of Staphylococcus aureus and a virulence factor in multiple models of infection (Kiedrowski et al., 2014). Given the ion-dependence of the MNase, we developed an MST assay to test the binding of different mono- and divalent ions to MNase. As the ions do not display any fluorescence, it was possible to perform a truly label-free, in solution MST assay by using the MNase tryptophan fluorescence for MST detection (“label-free” assay).
Results: Ion binding to S. aureus MNase
As expected from its Ca2+-dependence, MNase showed a strong preference towards divalent cations such as Ca2+, Mg2+, Mn2+, and Zn2+ with KD-values in the low millimolar concentration range (KD: 3.6 ± 0.9 mM, 9.9 ± 5.4 mM, 0.9 ± 0.4 mM and 0.2 ± 0.1 mM). In contrast, no interaction of MNase and Na+ was detectable (Figure 2). This assay highlights not only the potential of MST to study membrane-associated proteins but also to detect binding of very small ions to large proteins; a task that is not feasible with most other biophysical methods (or requires much more effort).
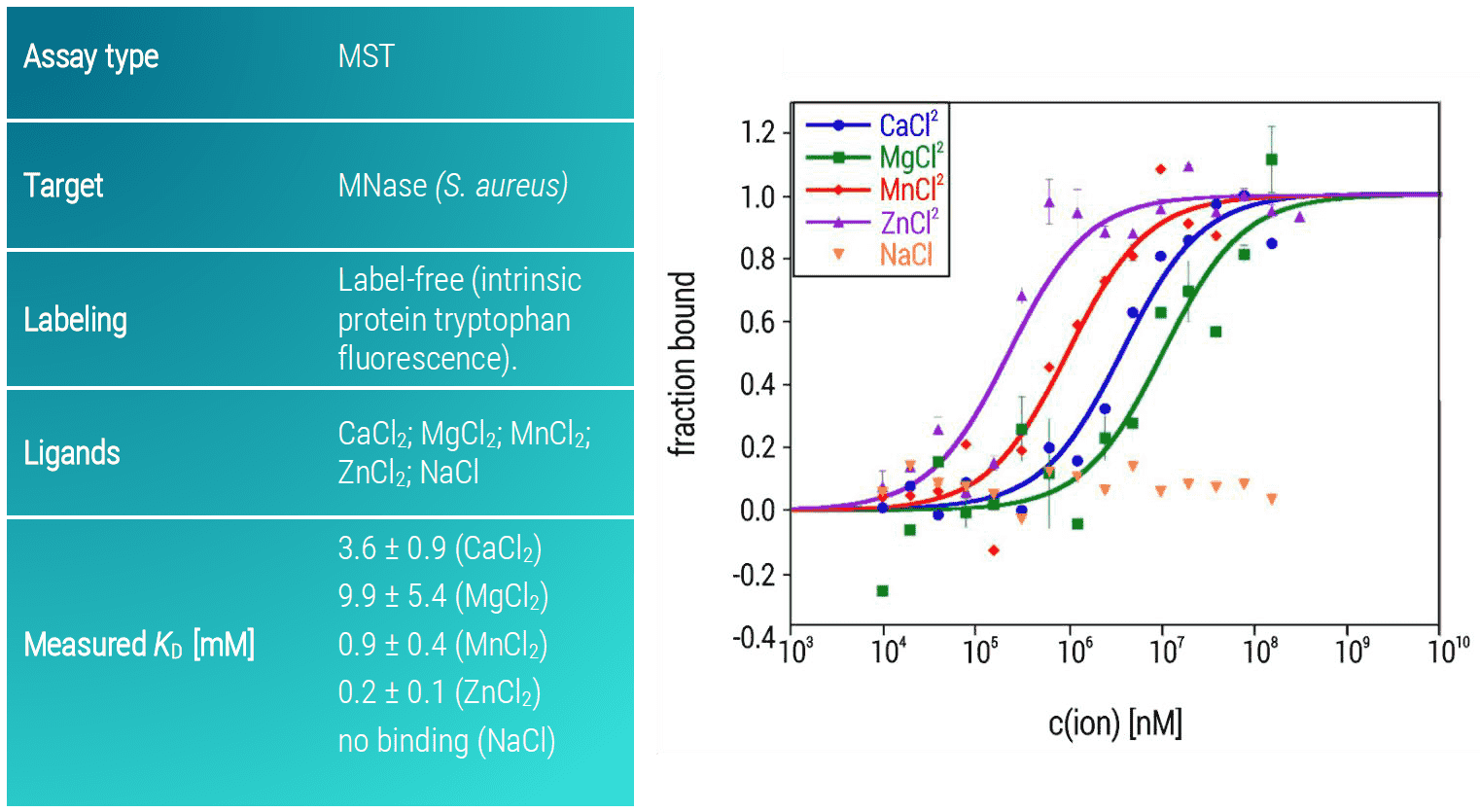
Figure 2: MST binding isotherms of MNase and various mono- and divalent ions. Data are the mean and standard deviation of two independent experiments and are fitted to a KD‑binding model under a 1:1 binding situation.
Conclusion: 2bind MST assays for measuring binding of ions to proteins
MST assays are very versatile, because they can easily accommodate different buffers, detergents, as well as target and ligand types (proteins, nucleic acids, small molecules, ions). Here, we successfully established an MST assay that allows an analysis of the binding affinity of enzymes and their ionic cofactors in a fast and precise way.
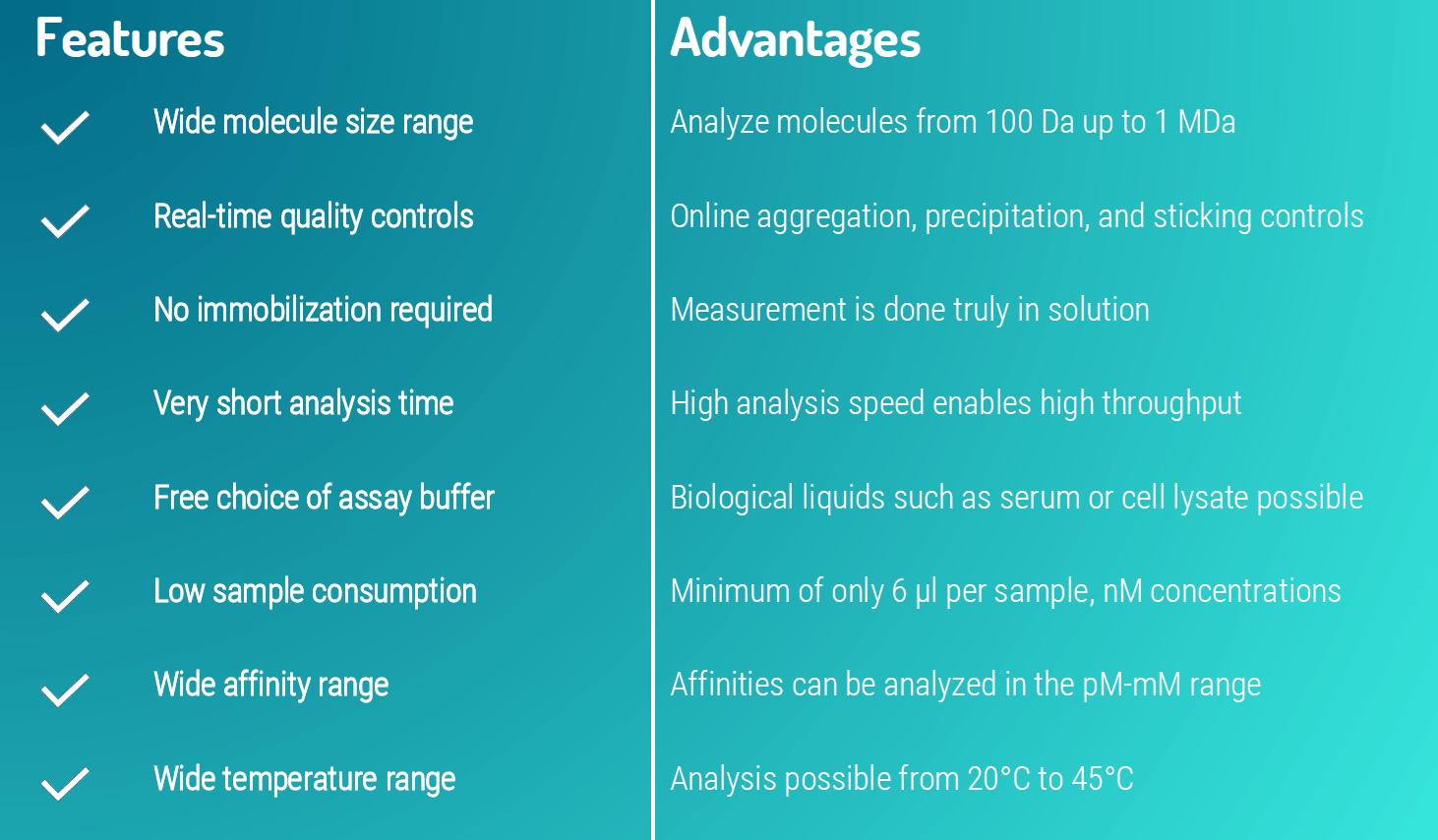
Since MST features a very low sample consumption (only 6 µl per sample is required, with concentrations of fluorescent target down to 0.5 nM), a wide molecular size range (100 Da- 1 MDa), a very short analysis time, and the strongpoint that measurements require no target immobilization, MST should be your method of choice for studying challenging targets such as membrane proteins. Moreover, MST works with virtually every available detergent, which can be important for solubilization of membrane proteins.
Click here for a full list of services for protein interaction analysis. If you like to directly contact us for any kind of service, use the contact form below.
Literature
Baaske et al. Optical thermophoresis for quantifying the buffer dependence of aptamer binding. Angew. Chem. Int. Ed Engl. 2010; 49(15): 2238-2241
Duhr & Braun. Why molecules move along a temperature gradient. Porc. Natl. Acad. Sci. USA 2006; 103(52): 19678-19682.
Gupta, Duhr & Baaske. MicroScale Thermophoresis (MST). Encyclopedia of Biophysics. 2018; 1–5
Kiedrowski et al. Staphylococcus aureus Nuc2 Is a Functional, Surface-Attached Extracellular Nuclease. PLoS One. 2014: 9(4): e95574.
Overington et al. How many drug targets are there? Nat. Rev. Drug. Discov. 2006; 5:993–996.
Rask-Andersen et al. Trends in the exploitation of novel drug targets. Nat. Rev. Drug. Discov. 2011; 10:579–590.
