Analysis of Protein-DNA and Protein-RNA interactions using Biophysics (MST)
Clemens Entzian, Corinna Kuttenberger, and Thomas Schubert
2bind GmbH, Regensburg, Germany
MST Technology
MicroScale Thermophoresis (MST) is an optical fluorescent method. It records the changes in fluorescence of a target molecule as a function of temperature and the concentration of the cognate ligand molecule. Thus, in MST, the target must be fluorescent and the ligand must not be fluorescent in the same wavelength range as the target. The fluorescence changes are the result of two effects:
First, molecules exhibit directed movement along temperature gradients in solution, an effect called thermophoresis (Duhr & Braun, 2006). Such thermophoretic movement of a molecule is characterized by its size (hydrodynamic radius), its surface charge, and its hydration shell. All three parameters can be affected by binding of a ligand molecule. Thus, a concentration-dependent titration of the target molecule’s ligand induces changes in the thermophoretic movement. These changes translate in quantifiable, spatial fluorescence changes, which are easily tracked optically via the target fluorescence.
Second, fluorescence is a function of the temperature. Thus, the fluorescence of the target molecule varies with the temperature (Baaske et al., 2010). This temperature-dependence is additionally affected by the local molecular surroundings of the fluorophore. Thus, binding of a ligand molecule to the fluorescent target can change the chemical environment of the target fluorophore and thus the overall detected temperature-dependence of the fluorescence. This effect is known as TRIC (temperature-related intensity change; Gupta, Duhr & Baaske, 2018).
The thermophoresis and TRIC signals are additive and thus both contribute to the high sensitivity and robustness of MST measurements towards molecular binding events of all kinds. Thus, MST can be used for determining the affinity and binding strength of almost any kind of molecular interaction with very low sample consumption and very high sensitivity.
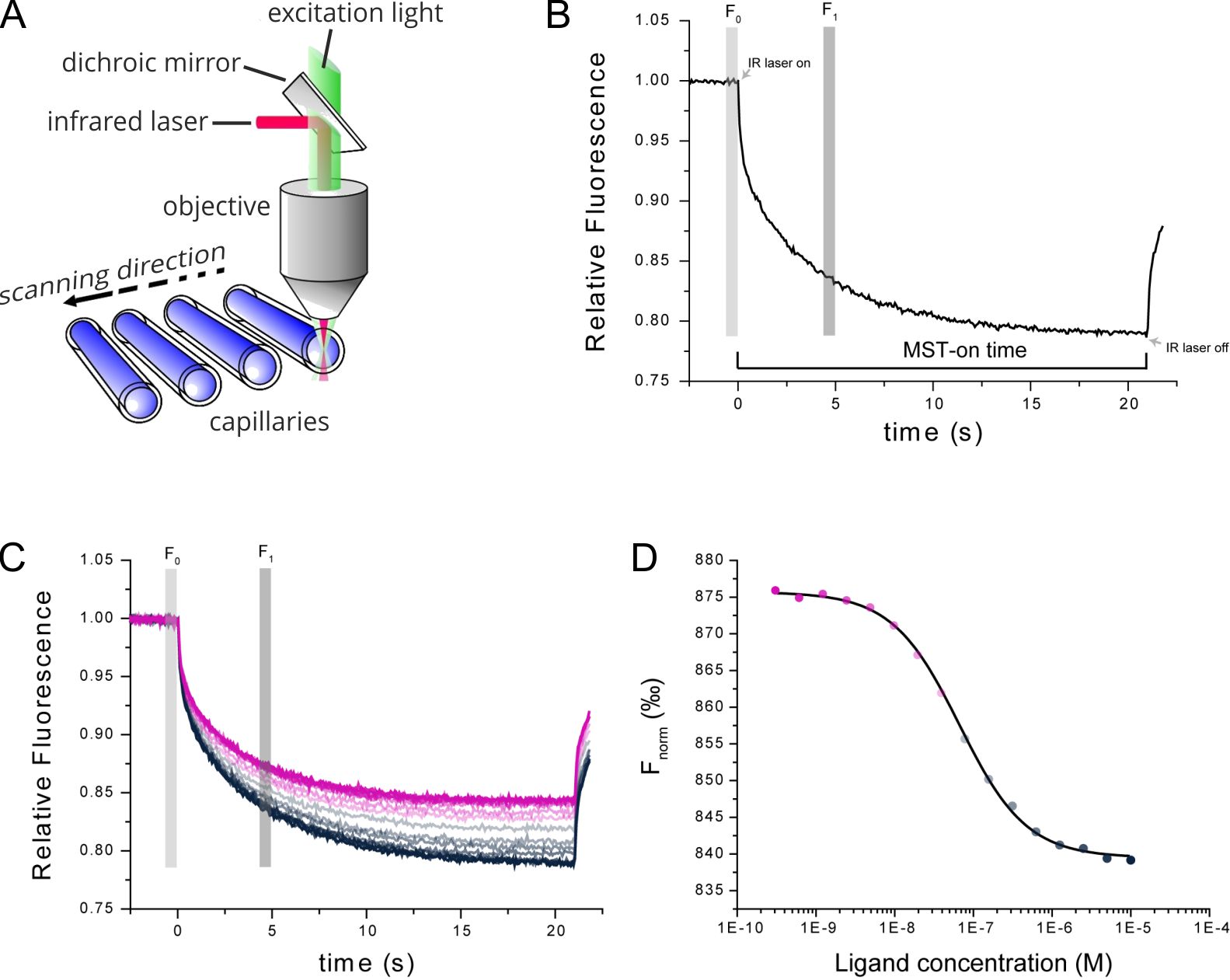
Figure 1. Technical MST setup. (A) MST measurements take place in small glass capillaries. Infrared and fluorescence lasers are used for generation of the MST effect and sample tracking. (B) TRIC and thermophoresis together account for a time-dependent change in fluorescence upon infrared-heating of the sample capillaries. (C) Multiple MST traces are recorded for different mixture ratios of target and ligand molecules. (D) Dose-response analysis of the MST traces allows for determination of the steady-state affinity of the target-ligand interaction.
Introduction: Interactions between protein and DNA / RNA
Interactions of proteins and nucleic acids are reported to play a pivotal role in epigenetic networks, especially in the chromatin decondensation processes (Schubert et al., 2012). Dysregulated epigenetic networks represent a major reason for disease development and thus analysis of protein-nucleic acid interactions can deliver a multitude of new drug targets. It is often complicated to access the basic binding parameters such as steady-state affinity of nucleic acid fragments using biophysical methods.
Here, we demonstrate how MST can be used to determine the affinity of the epigenetic effector protein Df31 to single stranded (ss) RNA-, ssDNA-, or double stranded (ds) DNA fragments. The nucleic acid fragments shared the exact same sequence and length and differed only in their ss/ds “nature”.
For this, a simple yet robust MST assay was developed, in which the nucleic acid fragments were used as the constant target interaction partner, labeled with a Cy5 fluorescent dye for optical tracking during the measurements. When working with nucleic acids this labeling strategy often makes sense, because the nucleic acids can by directly synthesized with a fluorescent dye attached to their 5’ or 3’ ends. The Df31 protein was used as the ligand and titrated in 16 steps across a concentration range from 800 µM down to 350 nM.
Results
We observed that Df31 specifically binds the ssRNA fragment with a KD value of 24 ± 3.1 μM and fails to recognize the single-stranded or double-stranded fragments (Figure 2). Importantly, the whole nucleic acid profiling just required single-digit nM concentrations of the nucleic acids. This can be of high importance when working with longer and expensive nucleic acids (e.g. with chemically modified bases). Although the used concentrations of the DF31 were quite high – in order to cover its affinity range with a full dose-response curve – the used volumes were very low. For each experiment, only 5 µL of the highest concentration solution were required. Thus, the whole profiling only required 15 µL of the Df31 protein.
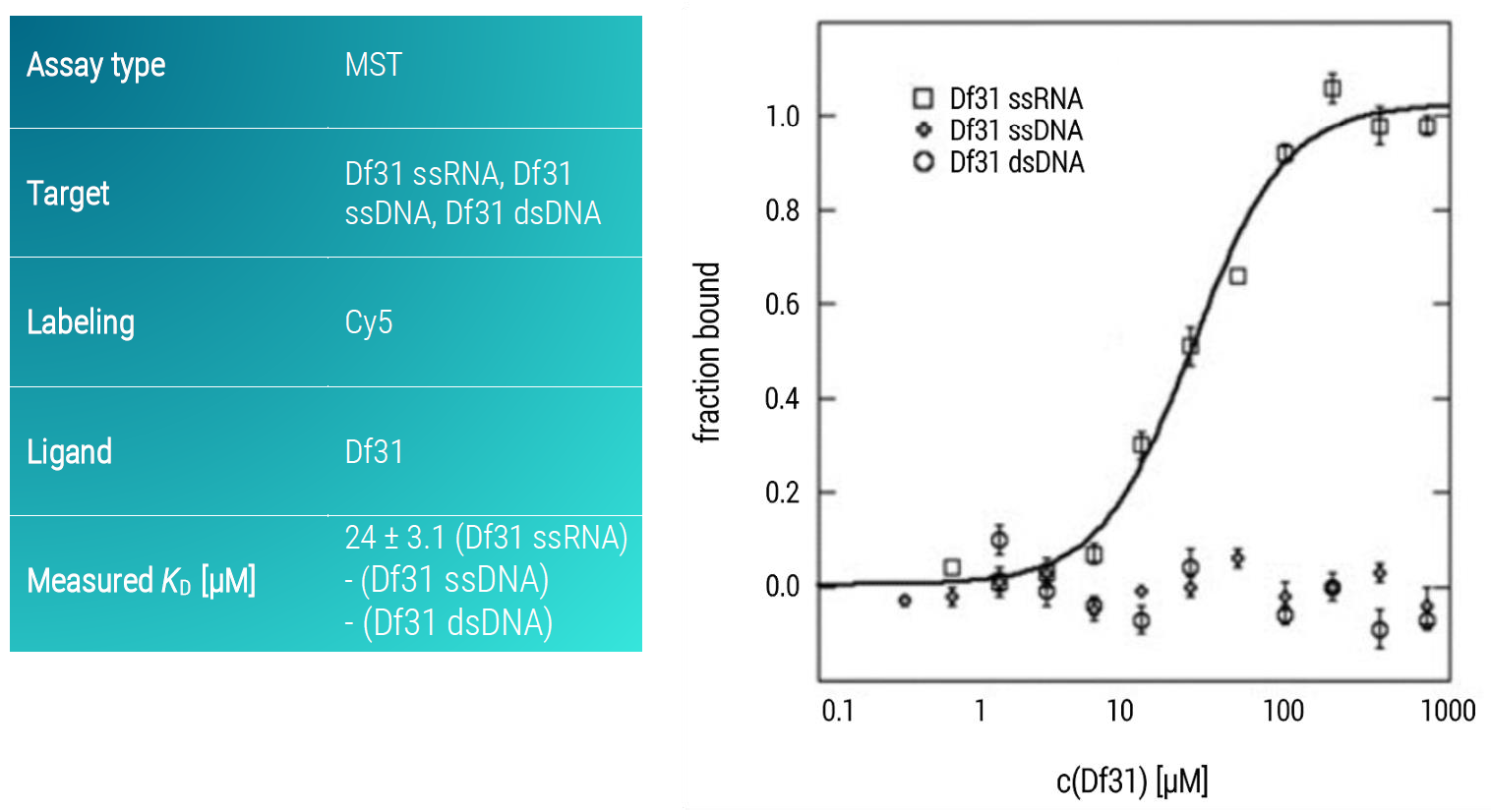
Figure 2: MST dose-response curves of fluorescently labeled ssRNA, ssDNA, and dsDNA of the same sequence and length (39 nucleotides) to Df31. The SsRNA-Df31dose-response curve was fitted to the Hill equation, data represent the mean ± standard deviation from three technical replicates.
Conclusion: 2bind MST assays for measuring binding of DNA/RNA to proteins
In this application note, we demonstrated that our versatile MST assay is capable of measuring interactions between nucleic acids and binding proteins in a fast and robust manner as well as with minimal sample consumption. Similar assays can be readily developed for other questions of nucleic acid-protein binding.
Literature
Baaske et al. Optical thermophoresis for quantifying the buffer dependence of aptamer binding. Angew. Chem. Int. Ed Engl. 2010; 49(15): 2238-2241
Duhr & Braun. Why molecules move along a temperature gradient. Porc. Natl. Acad. Sci. USA 2006; 103(52): 19678-19682.
Gupta, Duhr & Baaske. MicroScale Thermophoresis (MST). Encyclopedia of Biophysics. 2018; 1–5
Schubert et al. Df31 Protein and snoRNAs Maintain Accessible Higher-Order Structures of Chromatin Mol. Cell. 2012; 48(3): 434-444
