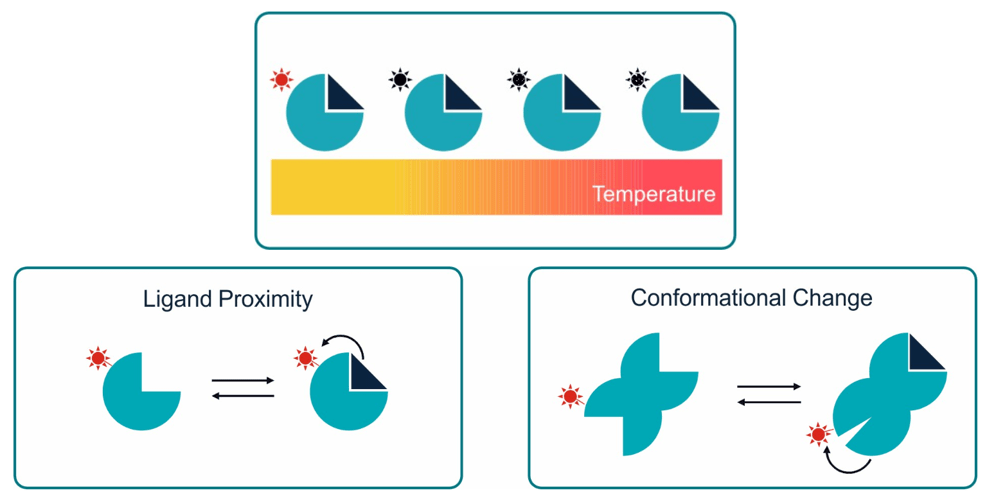
TRIC is based on the temperature-dependent properties of fluorescent molecules. When illuminated by a laser, these molecules emit light, and the intensity of this light changes with the environment of the molecules.
To utilize TRIC for measuring biomolecular interactions, a fluorescent dye is attached to the target molecule, e.g. a protein, and this now fluorescent target is brought into a microscopic temperature gradient (induced by a laser). The dye fluorescence on a target molecule bound to its interaction partner (e.g. a small molecule or another protein) experiences a different thermal effect compared to when its present of a “free” target, thus influencing the emitted light.
The choice of dye, properties of the target and ligand molecules, and successful labeling of the target significantly impact the sensitivity and accuracy of TRIC/MST measurements. 2bind's team of experts has extensive experience in selecting the right dyes for diverse biomolecules, along with performing efficient and reliable labeling procedures. We ensure the best possible detection limits for your specific interaction, eliminating the need for in-house labeling efforts.
KD
Equilibrium dissociation constant. Can be obtained by kinetic or classical equilibrium binding analysis. Provides information about the strength but not the dynamics of an interaction.
ΔH
Binding enthalpy. KD values at different temperatures can be used to obtain the binding enthalpy of an interaction via van't-Hoff-plots.
Ligand scouting
Fast single-dose screening for qualitative (Yes/No) binding.
Competition Assays
Determination of competition of several ligands for a selected binding site on a target.
Complex Inhibition Assays
Determination of binding site blocking by a ligand and prevention of binding of a natural interaction partner.
Stoichiometry Assays
Determination of equilibrium binding stoichiometry of target-ligand complex formation.
From library screening to competition assays: Accelerate Your Research with 2bind’s TRIC/MST Expertise
Whether you’re in the early stages of library screening, small molecule, fragment- or RNA-targeted drug discovery, seeking novel protein-protein interaction inhibitors like PROTACs, or optimizing DNA/RNA aptamers, 2bind’s TRIC/MST services offer the perfect solution. Our team’s vast experience translates into optimized workflows, tailored experimental designs, and in-depth data analysis, all geared towards achieving your specific goals.
TRIC, at its core, relies on the temperature-dependent sensitivity of fluorescent molecules. When illuminated by a laser, these molecules emit light, and their intensity can change based on their surrounding environment. To utilize TRIC for measuring biomolecular interactions, a fluorescent dye is attached to the target molecule, e.g. a protein, and this fluorescently labeled target is brought into a microscopic temperature gradient. Target molecules bound to an interaction partner or small ligand experience a different thermal environment compared to their free target state. This difference in environment affects the fluorescent properties of the attached dye, influencing its light emission. By measuring these changes in intensity as the temperature rises, TRIC reveals crucial information about the binding interaction.
Binding of a ligand to the fluorescently labeled target molecule usually either directly induces changes in the surrounding environment of the attached dye (i.e. close-proximity binding) or translates to minute conformational changes that propagate to the surroundings of the attached dye.
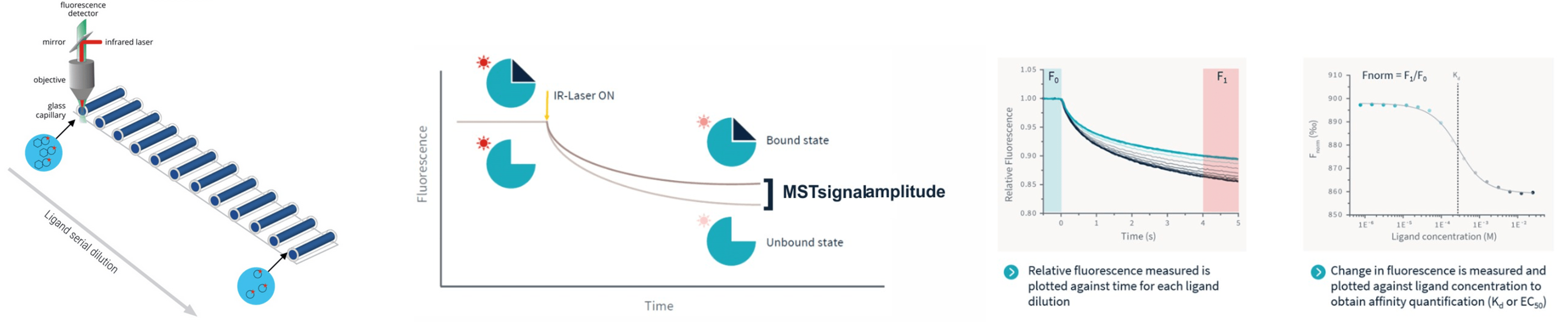
TRIC/MST measurements are done in a glass capillary setup. An infrared laser is used to generate a minute temperature gradient while an LED excites the fluorescent dye on the target molecule. A series of capillaries contain a mix of fluorescent target molecule and increasing concentrations of non-fluorescent binding partner. The sample fluorescence in each capillary is then measured over time to determined the baseline fluorescence. Then, the infrared laser is turned on. Again, the fluorescence is monitored over time and any changes in fluorescence due to the TRIC effect are recorded as so called TRIC-traces or MST-traced. From these, the “Fnorm” values are calculated (fluorescence in region F1 divided by fluorescence in region F0). In case of a productive interaction between target and ligand, the Fnorm values are dose-dependent and can be well described by the law of mass action. A plot of Fnorm against ligand concentration then returns the dissociation constant KD of the interaction.
TRIC/MST works best with fluorescent dyes in the “red” wavelength spectrum, i.e. dyes with excitation maxima around 650 nm and emission maxima around 670 nm. Commonly, specialized TRIC-dyes are used, which maximize the obtainable TRIC response by being very temperature sensitive. However, also more standard dyes like Cy5 work well with TRIC/MST, for example when used for DNA- or RNA-labeling. 2bind also uses specialized TRIC/MST instruments that allow detection of other fluorophores like Cy3, Alexa647, GFP or other “green” fluorescent dyes.
Yes, TRIC can also be measured with protein-intrinsic Tryptophan fluorescence. However, since Tryptophan fluorescence has not “evolved” for maximum TRIC response, the obtainable signals are usually smaller and careful assay development and controlling of assay conditions is necessary.
No, for protein targets, 2bind performs all required steps for fluorescence labeling of the target. Peptides, DNA, or RNA targets are often synthesized directly with a selected fluorescent dye attached.
The most common labeling methods are covalent NHS- or Maleimide-chemistry reactions. These reactions attach fluorescent dyes directly to lysine or cysteine residues of the target protein, respectively. This is a routine part of the assay process at 2bind. The advantage of these covalent dye labels are the option to use very low (single-digit nM) concentrations of the labeled target protein and therefore determine even sub-nM KD values accurately. An alternative to covalent labelings are site-specific labelings at a His-tag, SNAP-tag, or biotinylated Avi-tag of the target protein.
TRIC is perfect for any bi-molecular interactions between any kind of biomolecule: proteins, peptides, DNA, RNA, aptamers, small molecules, fragments, lipids, carbohydrates, ions, or even large artificial or virus(like) particles. Since the size of the interaction partners is not the dominant driving force behind the TRIC measurement, almost all these classes of biomolecules can be tested with the same accuracy and precision.
2bind offers the following assay types with TRIC/MST: Hight-throughput interaction screenings, Steady-state binding affinity assays, Steady-state binding affinity assay in biological liquids, Complex formation-inhibition assays, Competition assays, Binding assays with multiple binding partners.
TRIC/MST offers many advantages compared to methods for determining the affinity of a molecular interaction:
– Low sample consumption: As little as 50 µM of protein material is enough to perform one labeling reaction. The output of this one labeling reaction is often enough to test close to 100 ligands.
– Free choice of assay buffers: No non-compatible buffer substances. Also, biological liquids are possible such as serum or cell lysate.
– Very short analysis time allows for high-throughput binding screening.
– Online quality controls: Detection of aggregation, precipitation, and sticking of the target molecule and the option to quickly change buffer conditions.
– No surface-immobilization required: Measurement is done truly in solution.
– Wide concentration range: Affinities can be analyzed in the pM-mM range.
– Wide molecule size range: Ligands can range from ions to MDa particles.
TRIC/MST determines the equilibrium, steady-state binding affinity of two molecules (KD). Therefore, samples are analyzed after the binding reaction reaches chemical equilibrium. Measurement of kinetics is only possible if the equilibrium is not reached quickly. The thermodynamics of an interaction can be investigated using the van’t Hoff method. However, for in-depth kinetic analysis of molecular interactions other technologies like GCI or BLI are excellent. For thermodynamics, an ITC is preferred.
Yes, given that the association and reaction kinetics of the covalent binding ligands is not too fast (seconds-scale). If this is the case, a time-series of ligand dilution series can be measured with TRIC to shed light on the covalent binding parameters of a ligand. However, possible temperature effects from the infrared TRIC-laser must be taken into account. Therefore, for covalent binding analysis, Spectral Shift is usually recommended.
Yes, due to its inherently robust physical principle and technical readout (specific fluorescent dye on the target molecule), TRIC/MST can be measured even in 100% complex bioliquids like serum, plasma, cell lysate, urine, saliva, sea water, etc.