2bind’s deep expertise in Dynamic Light Scattering (DLS) will transform your biologics and therapeutics development workflows. The DLS technology rapidly measures the size and colloidal stability of proteins, biomolecules, and compounds. Our customized solutions for biologics development (proprietary buffer screens FORMOscreen® and KOMBOscreen®, Design-of-Experiment methodology, a.k.a DoE) and drug discovery (small molecule solubility and aggregation screening) paired with expert data interpretation, ensure high-quality data and actionable results. Choose 2bind for faster turnaround times, superior sensitivity, and unparalleled support at every step.
Dynamic Light Scattering (DLS) is a powerful, non-destructive technique that illuminates the behavior of proteins, macromolecules, and nanoparticles in solution. DLS passes a laser beam through a sample and the biomolecules in the sample scatter the laser light. The resulting scattering pattern reveals essential insights into the molecules’ size, stability, homogeneity, and their tendency to aggregate.
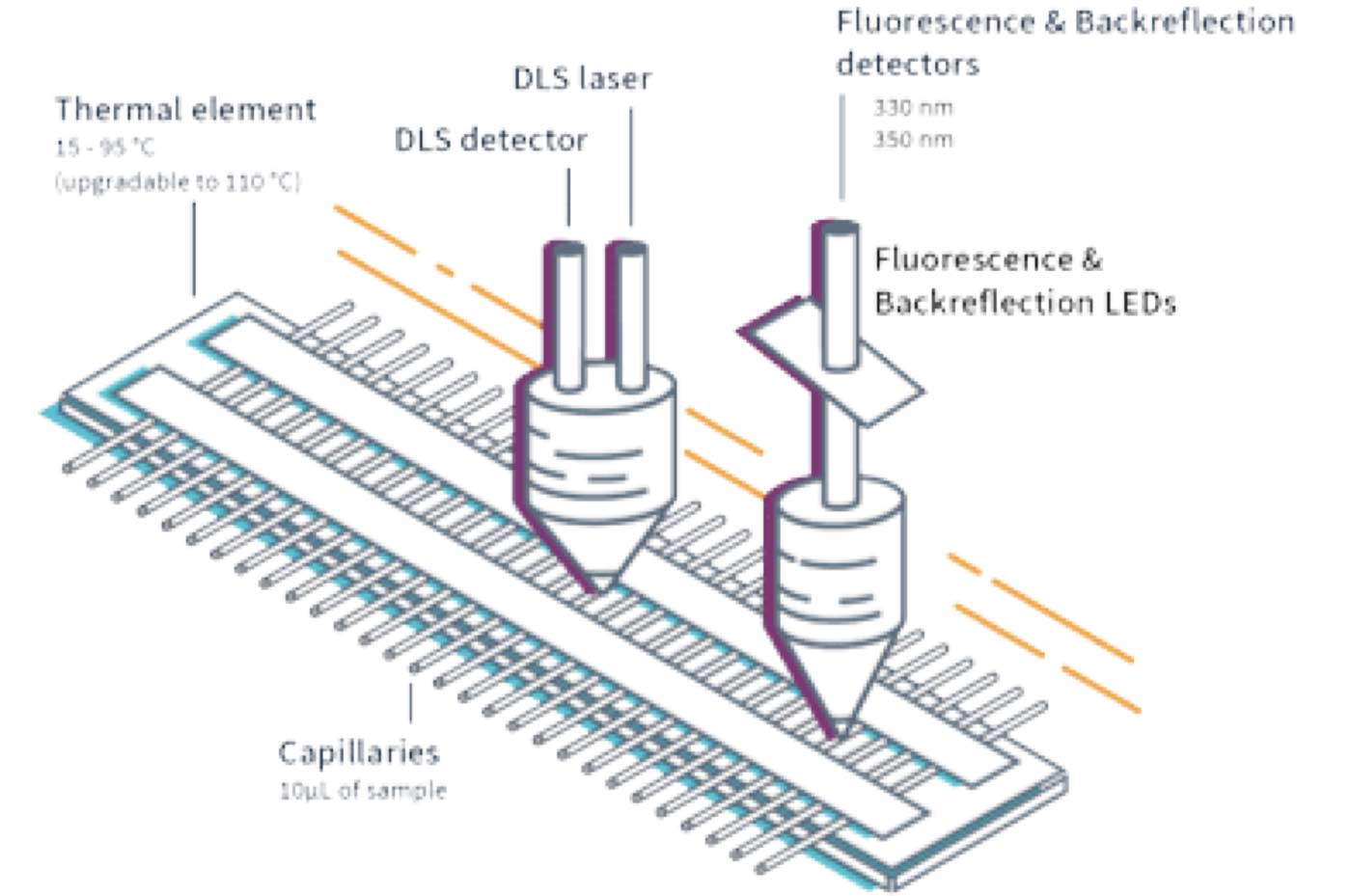
2bind leverages cutting-edge instruments like the Prometheus Panta and the Wyatt DynaPro Plate Reader III for highly precise DLS analysis. These tools offer remarkable sensitivity, enabling us to analyze challenging samples at low concentrations. Our experienced scientists translate the raw scattering data into knowledge about your biologic's behavior in different formulations, storage conditions, and temperatures.
rH
Hydrodynamic radius; the (average) size of the particles in solution
PDI
Polydispersity index; distribution of particle size populations
Tsize
Temperature at which average particle size begins to increase; from change of rH in cumulants fit)
Tscattering
Temperature at which particles begin to form scattering aggregates, from change of scattering intensity
kD
Diffusion-interaction parameter; shows whether molecular interactions are repulsive or attractive
D0
Theoretical diffusion constant at concentration = 0; determined from the kD analysis
B22
Particle interaction parameter; measure of the total protein-protein interactions in a solution
HMS content
Content of higher-molecular-weight species in small molecule solutions; used to derive aqueous solubility
Our sample preparation protocol carefully prevents any dust or particle contamination prior to measurement. In general, DLS is not limited by buffers or additives. However, it must be noted that every particle in the sample will scatter light. Thus, complex solutions might give background scattering signals that prevents successful DLS analysis.


DLS is a versatile technique that can be applied to all kinds of biological particles. Proteins are usually the main target for DLS analysis. Another major class of DLS experiments is development of solution characteristics, for example for biologics formulation development or cosmetics. Due to the fast analysis pace and high throughput, DLS is well suited for screening of buffers, buffer additives, or small molecule influence on protein stability and protein unfolding.
Normaly not. But our Prometheus Panta instrument series combines the ultra-high resolution of DLS with state-of-the art nanoDSF (nano-scale differential scanning fluorimetry) for simultaneous measurement of DLS and protein unfolding. It even allows for detection of multiple domain unfolding and aggregation events. Even changes in the DLS properties of individual protein domains during unfolding can be monitored, using the DLS regularization fit during thermal melting of a protein. Protein unfolding can be measured with the Wyatt DynaPro III using a technique called temperature ramping. The instrument measures the hydrodynamic radius of the protein at different temperatures, and the resulting data is used to generate a thermal denaturation curve. The unfolding temperature (Tm) can be calculated from this curve, providing insight into the protein’s stability and behavior under different conditions. This information is critical for the development of biopharmaceuticals and other protein-based products.
Yes, it is possible to observe the effect of ligand binding on the hydrodynamic radius by monitoring the cumulant radius of the target in the presence of increasing concentrations of the ligand.
Yes, it is possible to measure protein-protein interactions with DLS. For this, a continuous variation experiment can be performed, in which the concentrations of the two interaction partners are simultaneously and continuously varied and the mean particle size is determined. Upon binding, the largest particle size will be observed. Note that for this experiment to work, the differences in hydrodynamic radius of the single proteins must not exceed two-fold (or the molecular weight difference must not exceed 5-fold).
Our sample preparation protocol carefully prevents any dust or particle contamination prior to measurement. In general, DLS is not limited by buffers or additives. However, it must be noted that every particle in the sample will scatter light. Thus, complex solutions might give background scattering signals that prevents successful DLS analysis.