Reveal conformational changes induced by analyte binding or buffer conditions with switchSENSE analysis.
switchSENSE enables comparative studies of molecular friction, which reacts on size and conformation. Comparing the friction of a molecule in different defined situations provides provides insight into the reaction of a molecule onto its microenvironment. Small molecule analytes frequently impact the conformation of a protein upon binding, and the effects can be visualized and compared using the switchSENSE technology.
Besides comparing the molecules shape in different situations, switchSENSE can also be measured in real-time, opening new avenues for research and discovery.
switchSENSE is part of the powerful heliX® technology platform, which also offers FPS, kinFRET, and Real-Time Interaction Cytometry (RT-IC) for comprehensive biomolecular interaction analysis. At 2bind, our team of experienced scientists leverages the full potential of heliX® switchSENSE to deliver comprehensive solutions for your research needs and ensure accurate, reliable, and actionable results.
At the heart of switchSENSE lie DNA nanolevers tethered to a gold microelectrode. By applying an electrical potential, the orientation of these nanolevers can be precisely controlled, creating two distinct measurement modes, a dynamic mode and a static mode (for more information, please see FPS). In the dynamic switchSENSE mode, alternating potentials induce oscillations in the DNA nanolevers. These oscillations are affected by the overall size of the nanolevers, i.e. their overall molecular friction. Binding of molecules to the nanolevers, e.g. binding of an analyte to a target molecule that is immobilized on the nanolevers, leads to changes in the molecular friction and therefore in a detectable switchSENSE signal. Careful assay setup even allows for the detection of conformational changes induced by small molecule binding, e.g. ATP binding to a kinase.
Conformational change
Comparative measurement of the molecular friction. Reveals conformational changes, folding/unfolding, or agglomeration.
Relative sizing experiments
Relative changes in hydrodynamic radius can be detected and protein size (in kDa) can be estimated.
EC50/IC50
Dose-response data. Ligand concentration that gives half-maximal response or half-maximal activity.
Drug Discovery: Take deep dives into mechanism of action for small molecules and biologics, e.g. by investigating possible size or conformational changes upon target-ligand binding.
Basic Research: Investigate the fundamental mechanisms underlying protein-protein interactions, protein-DNA interactions, and conformational changes in biomolecules.
Quality Control: Develop robust and sensitive assays for monitoring the quality and stability of biopharmaceuticals.
The helix sensor chips comprise a gold microelectrode to which DNA nanolevers are tethered. Due to the inherent rigidity of DNA double helices and their negative charge, their orientation on the chip surface can be precisely controlled by applying an electrical potential to the electrode. Depending on the electrical potential, two distinct measurement modes are possible.
First, a static mode: A constant negative potential repels the nanolevers, maintaining them in an upright orientation. This mode utilizes fluorescence proximity sensing (FPS), where binding of an analyte to the ligand at the nanolever’s end causes a change in fluorescence intensity due to alterations in the dye’s microenvironment. Fluorescent dyes are attached to the distal ends of the DNA nanolevers. Single photon fluorescence counters facilitate highly sensitive detection of two distinct colors simultaneously, enabling multiplexed measurements or real-time referencing. Each electrode contains two measurement spots, allowing up to four different signals to be monitored. In the static (FPS) mode, the fluorescence intensity can decrease or increase upon binding of the analyte due to collisional quenching or anti-quenching effects and other processes. Fore more information on the static FPS mode see FPS.
Second, a dynamic mode: For the dynamic switchSENSE mode and oscillation of the surface-tethered DNA nanolevers is induced by alternating electric potentials applied to the gold microelectrode. The negatively charged backbones of double stranded DNA nanolevers are either attracted to or repelled from the sensor surface. This generates an oscillating orientation change of the DNA nanolevers, called switching. By observing the fluorescence emission of a fluorescent dye at the distal end of the DNA nanolevers, it is possible to assess the position of the DNA nanolevers relative to the gold surface. The intensity of the fluorescent light emitted by the dye reports on the distance to the gold surface due to a distance dependent radiation-free energy transfer to the gold. In other words, the closer the fluorophore is positioned to the quenching gold surface, the less light it emits. Thus, at positive (attractive) potentials, the DNA nanolevers align horizontally with the electrode’s surface, positioning the fluorescent dye close to the gold surface resulting in low fluorescence intensity. On reversing the electrode potential to a negative (repulsive) potential, the DNA nanolevers gradually move into an upright orientation, which is accompanied by a gradual increase of fluorescence intensity. Conformational changes are detectable by comparing to the initial state. Compaction results in reduced friction, whereas extension due to analyte binding or multimerization leads to increased friction.
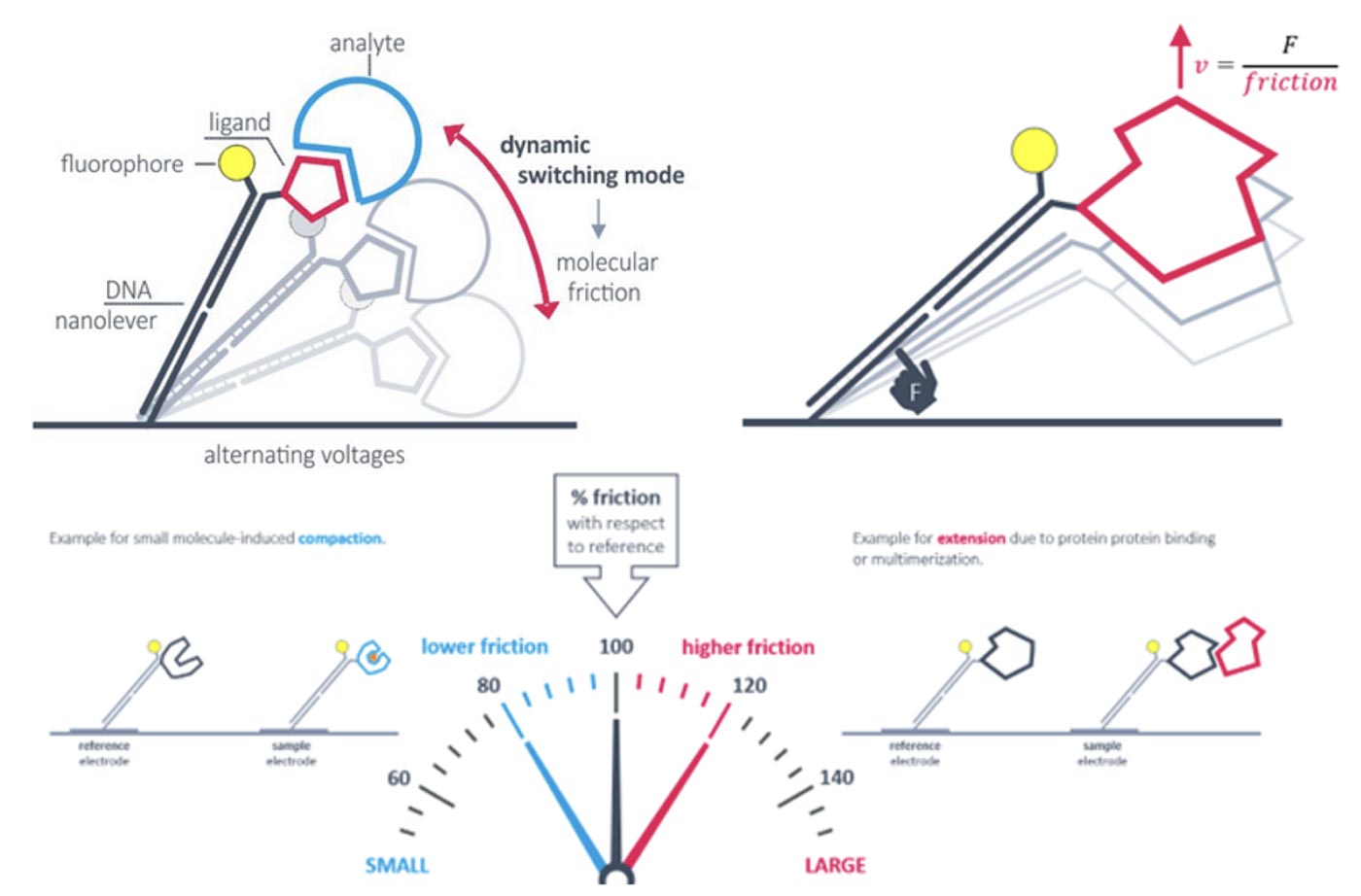
The dynamic switchSENSE measurement mode of the heliX® platform can be used to detect conformational changes because these will change the hydrodynamic friction of the attached molecule, which affects the switching speed.
Here is a list of typical applications of switchSENSE with the heliX® platform:
Small molecules or ions inducing conformational changes
Formation of multimeric protein complexes
Posttranslational modifications leading to conformational or structural changes
Structure-function relationship
Efficacy and mode of action of drugs
Key questions of assay design typically include the immobilization strategy. heliX® biosensor chips are equipped with single stranded anchor sequences and the most convenient way is the immobilization of target molecules via DNA hybridization. Different anchor sequences attached to electrode 1 and electrode 2 enable targeted immobilization to each spot. Typically, adapter strands carrying the fluorophore and ligand strands are prehybridized. Each electrode is equipped with a two-color detector and interactions of one analyte with up to four target molecules can be measured in parallel.
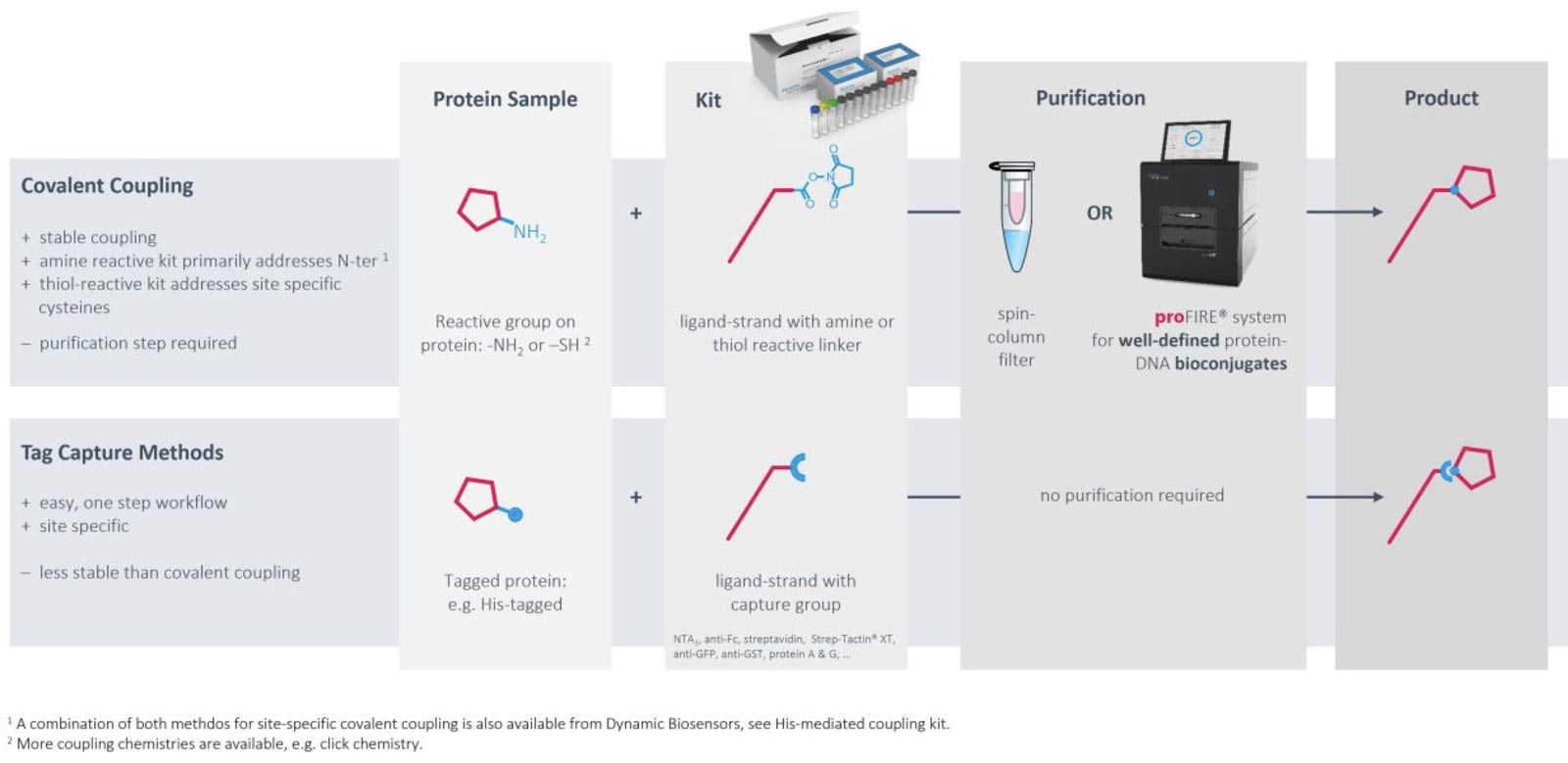
Immobilization via DNA hybridization is usually the method of choice to immobilize target DNA or RNA sequences. However, convenient, gentle and controlled conjugation methods as well as the proFIRE® purifier streamline the production of protein-DNA conjugates with high quality and purity, which can be easily implemented into the heliX® workflow. Ready-to-use kits for coupling to primary amines exist for molecules with usual pIs as well as for low pI molecules. If the target of interest contains a His-tag, coupling can be directed to amines in proximity of the tag. Alternatively, DNA strands can be coupled to thiol groups of free cysteines with high efficiencies and recovery rates.
As an alternative to chemical modifications of the targets of interest, tag-based capturing can be used. Methods and tools for capturing via His tags, biotin, twin-strep-tag, Fc parts and GFP have been developed and are ready to use.
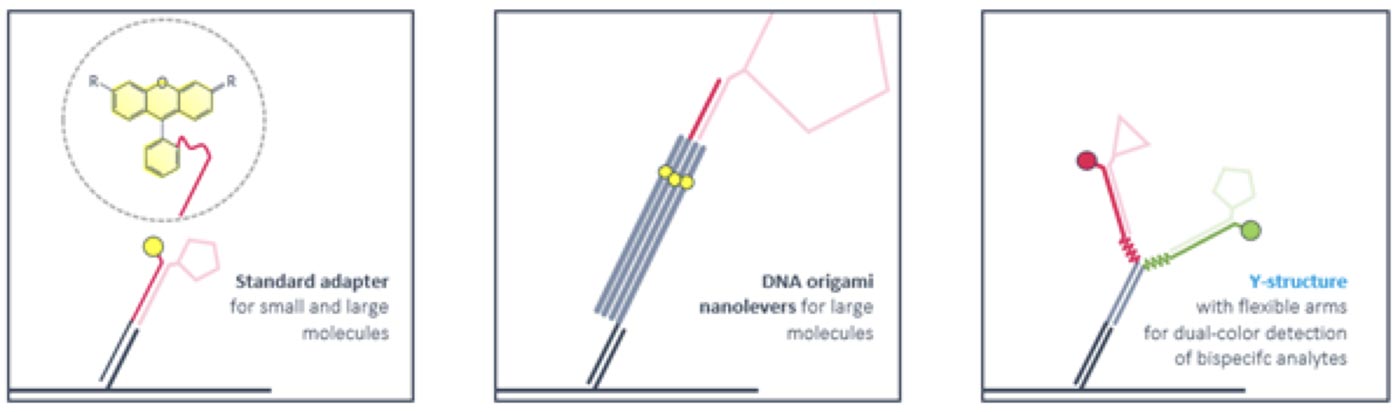
Ligand strands are compatible with standard adapters as well as with DNA nanostructures. Rigid DNA-origami nanolevers are recommended for sizing experiments with large molecules. Antibody-like Y-structures with flexible arms reduce the distance between attached ligands and enable bivalent binding of smaller bispecific molecules like PROTACS. Ternary complex formation and dimerization can be followed by time-resolved FRET.