FORMOscreen®: Buffers of Formulations of Developed Antibodies for Pre-Formulation of Biologics
Carina Schmid1, Corinna Popp1, Benjamin Zimmer1, David Peterhoff2, Ralf Wagner2, and Maximilian Plach1
1 2bind GmbH, Regensburg, Germany
2 Molecular Microbiology (Virology), Institute of Medical Microbiology and Hygiene, University of Regensburg
Features
| 96 buffer conditions from FDA- and EMA-approved formulations of therapeutic antibodies. | |
| Ready-to-use 96 x 0.2 mL buffer stock solutions (5x). | |
| Compatible with all major biophysical methods. | |
| Wide range of different buffer compositions: | |
|
pH: 4.6 – 8.0 (acetate, citrate, glycine, histidine, phosphate) Salts: 3 – 200 mM (potassium chloride, sodium chloride) Amino acids: 1 – 300 mM (glycine, glutamate, methionine, proline) Carbohydrates: 12 – 300 mM (maltose, mannitol, sorbitol, sucrose, trehalose) Detergents: 0 – 1.6% (polysorbate 20, polysorbate 80) |
|
| Purchase as a kit or use the 2bind analysis service (FORMOscreen® + nanoDSF) |
Introduction
Biologics-based medicines offer tremendous possibilities for personalized healthcare and are helping more and more patients with previously unmet medical needs, because of their targeted approach and high specificity. Not surprisingly, the global revenue from biologics is expected to increase to over 375 billion US dollars in 2022 (Statista, 2017). Among biologics, monoclonal antibodies (mAbs) captured the largest share in 2017 (Grand View Research, 2018) and are also technologically the leading segment (Walsh, 2018). As of 2018, over 80 monoclonal antibody drugs have been approved for clinical use by the FDA (Food and Drug Administration, USA) and the EMA (European Medicines Agency, EU). Key steps in the development of therapeutic antibodies are the optimization of their short- and long-term stability, as well as the definition of pre-formulation and final formulation conditions.
Pre-formulation development is usually a work-intensive and time-consuming process where many buffers have to be prepared, differing systematically in pH as well as salts, sugars, additives, and detergents. A quicker alternative is the 2bind FORMOscreen®: A collection of 96 proven, ready-to-use buffers from Formulations of Developed Antibodies approved by the FDA or EMA. The FDA-plate is compatible with your biophysical or biochemical read-out of choice (e.g. DSF, nanoDSF, DSC, DLS, SLS, LC-MS, HPLC-SEC, ELISA, etc,), allowing for quick and easy analysis of important antibody parameters: Chemical, thermal, colloidal, and conformational stability, long-term storage stability, forced-degradation resistance, as well as biochemical activity and antigen-binding. Importantly, all buffer conditions provided with the FORMOscreen® have already proven to be beneficial for use with antibodies. Just the same, all buffers have already been approved for use in antibody formulations by the FDA and EMA.
In this example study, we used the FORMOscreen® in combination with a high-throughput nanoDSF approach to find optimal stability and storage conditions for two monoclonal HIV-neutralizing antibodies. Additionally, their associated antigen, an HIV-1 envelope trimer was analyzed in order to highlight the potential of the FORMOscreen® for pre-formulation of non-Ig proteins.
Antibody Pre-Formulation
PGT145 antibody
Thermal stability
The thermal unfolding profile of the antibody PGT145 (Walker et al. 2011, Lee et al. 2017) shows a single unfolding transition in its storage buffer (citrate pH 5.9, 200 mM NaCl) with a melting temperature (Tm) of 63.7 °C (Fig. 1A). When the same antibody is measured in the 96 FORMOscreen® conditions (Fig. 1B), a large range of different melting temperatures are observed (46.7 – 68.4 °C). Thus, the antibody shows a pronounced variability in its thermal stability of 21.7 °C with respect to the buffer conditions.
The screening suggests that the optimal buffer for PGT145 thermal stability is a citrate buffer at pH 6.5 with 0.4 mg/mL Polysorbate-80, and 80 mg/mL Sucrose. In comparison to the storage buffer, this buffer results in a shift of Tm of + 4.7 °C. This may not seem much; but PGT145 is commercially available and already stability optimized. But the FORMOscreen® analysis could still improve its stability. In contrast, buffers containing sodium acetate, glutamate, proline, and in general buffers with a low pH should be avoided in order to not reduce the antibody thermal stability by up to 17 °C.
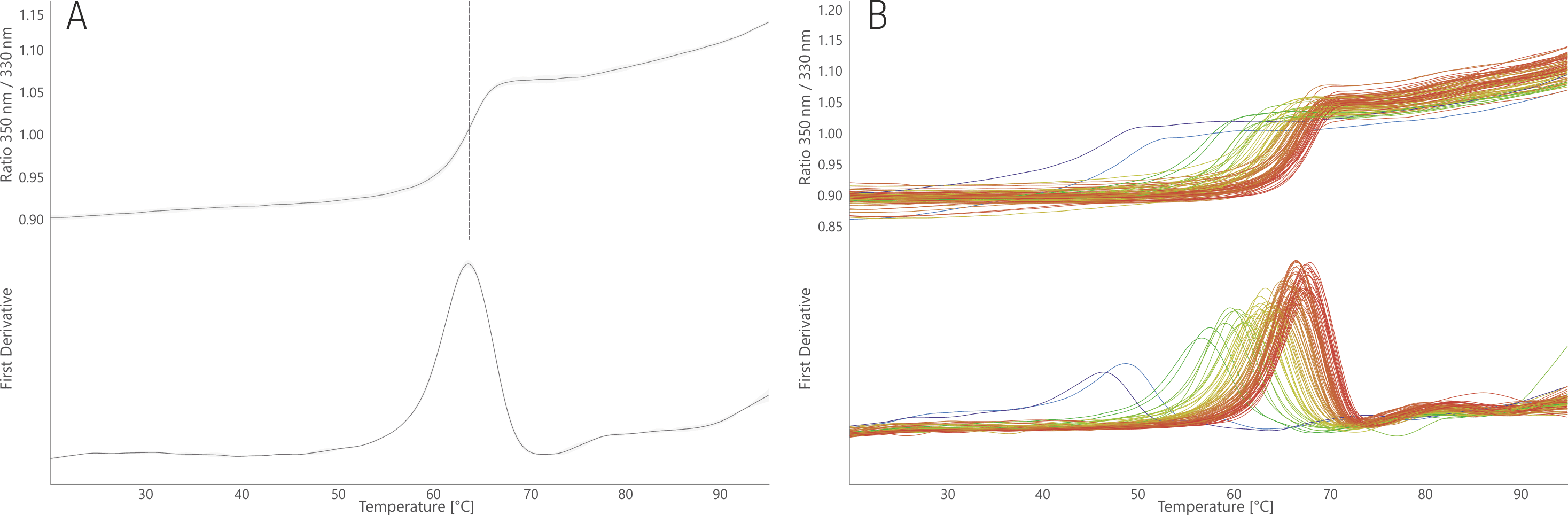
Figure 1. NanoDSF thermal melting of PGT145 stock condition (A) and in the 96 FORMOscreen® conditions (B). The upper panels show the raw fluorescence signal (350/330 nm ratio); the lower panels show the first derivative curves. Unfolding traces are colored from low (blue) to high (red) melting temperatures.
Aggregation
In its stock buffer, PGT145 aggregated at 79.2°C (Fig. 2A). In the FORMOscreen® conditions, its aggregation temperature ranged from 68.0 to 83.2 °C (Fig. 2B). Thus, a shift of almost 4°C was possible in the “best” buffer for protein aggregation. Interestingly, this buffer was not equally “good” for thermal stability. In fact, many buffers that induce high thermal stability (red curves in Fig. 1B), lead to relatively low aggregation temperatures (red curves in Fig. 2B). Thus, the FORMOscreen® analysis of this antibody reveals intricate differences between thermal stability and aggregation propensity.
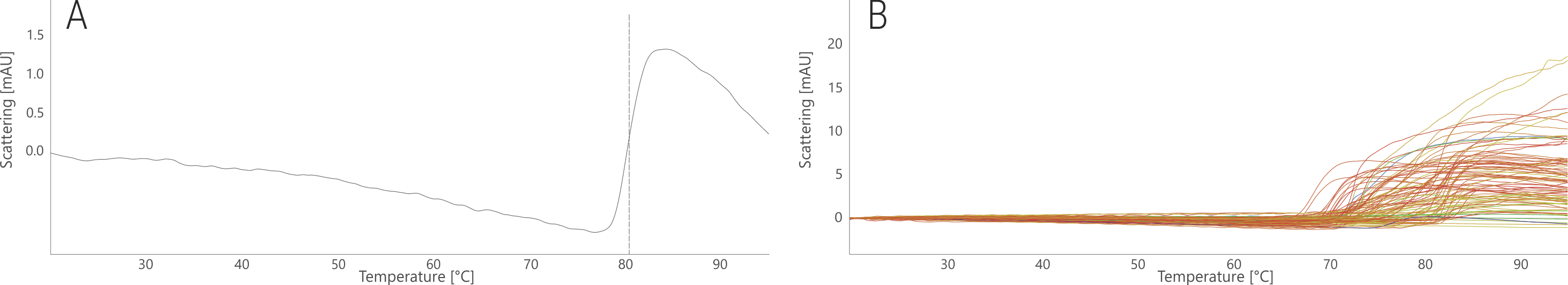
Figure 2. NanoDSF thermal aggregation analysis of PGT145 stock condition (A) and in the 96 FORMOscreen® conditions (B). Traces are colored based on the melting temperatures shown in Fig. 1.
17b antibody
Thermal stability
The thermal unfolding profile of the antibody 17b shows two distinct unfolding transitions at 65.3 and 76.6°C (Fig. 3A). The two transitions result from unfolding of individual antibody domains (Thali et al. 1993, Kwong et al. 1998). In most FORMOscreen® conditions this dual unfolding profile is retained (Fig. 3B). The highest shift observed for transition one was +10.4 °C and for transition two +3 °C. Thus, the stability of certain domains reacts more sensitive to buffer changes than that of other domains. In some conditions, mostly containing acetic acid as the buffer substance, only one unfolding transition was observed between 72.4 and 75.7 °C. This demonstrates that under certain conditions, the structural stability of the antibody domains can align.
Depending on the precise requirement and further use, an optimal buffer for 17b could be sodium acetate buffer at pH 6.8. This buffer results in a shift of + 4.4 °C for the first and of + 3.0 °C for the second unfolding transition. Buffers containing arginine and poloxamer or buffers with low pH levels should be avoided in order to not reduce the antibody thermal stability by up to ‑ 8.1 and ‑ 3.0 °C, respectively.
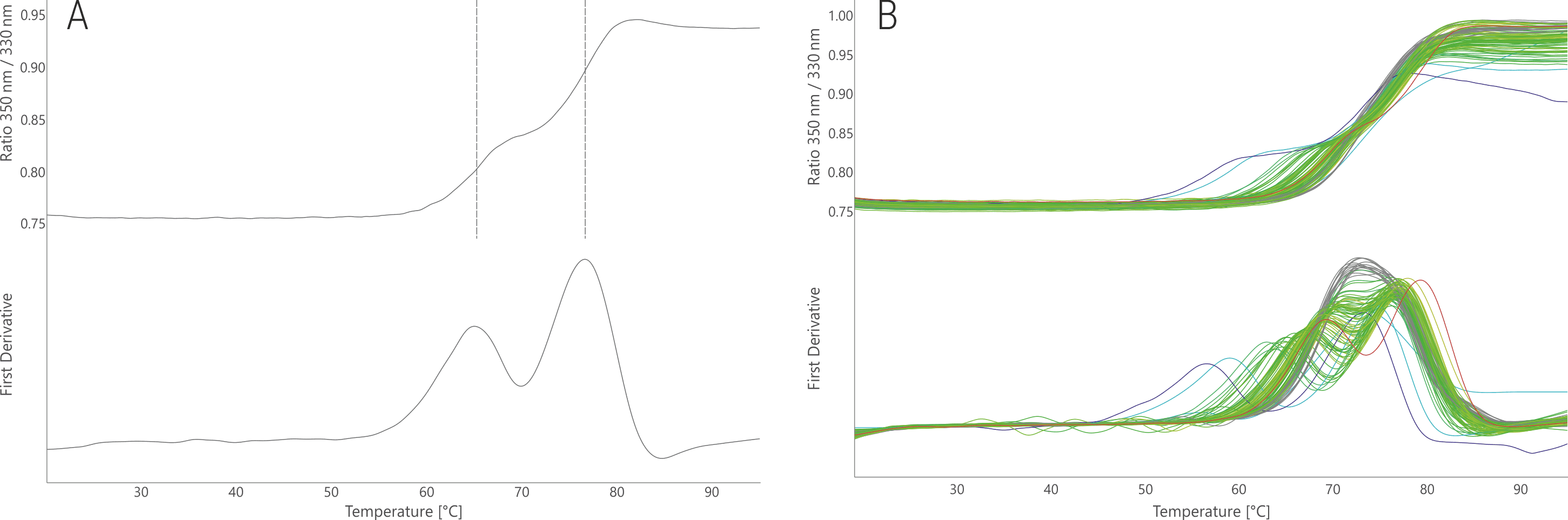
Figure 3. NanoDSF thermal melting of 17b stock condition (A) and in the 96 FORMOscreen® conditions (B). The upper panels show the raw fluorescence signal (350/330 nm ratio); the lower panels show the first derivative curves. Unfolding traces are colored from low (blue) to high (red) melting temperatures with respect to the first unfolding transition only.
Aggregation
In its stock buffer, 17b aggregated at 80.7 °C (Fig. 4A). The FORMOscreen® conditions mostly resulted in lower aggregation temperatures (- 18.3 °C at most); the highest positive shift observed was +0.3 °C (Fig. 4B). Although no significant stabilization with respect to aggregation temperature could be achieved, the FORMOscreen® analysis of 17b revealed which buffer conditions must be avoided in order to not increase antibody aggregation. In contrast to PGT145, there was a clear correlation between conditions with high/low thermal stability and high/low aggregation onset temperatures.
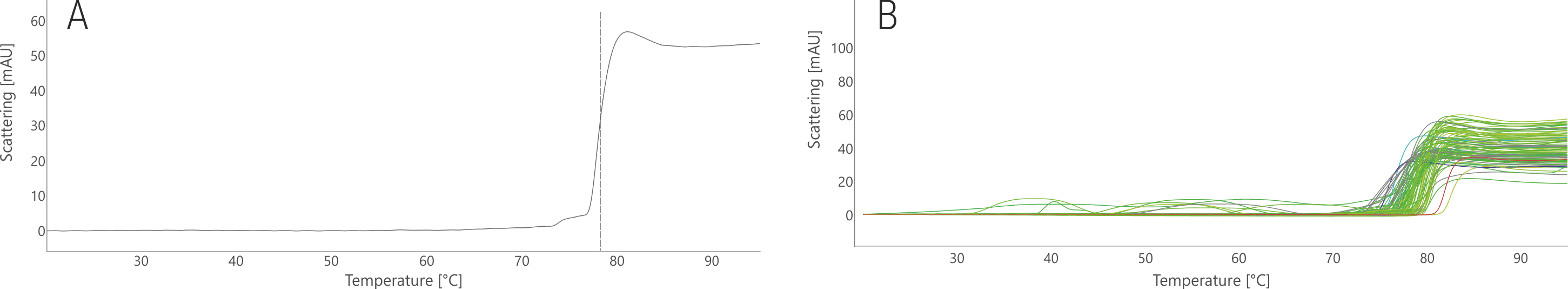
Figure 4. NanoDSF thermal aggregation analysis of 17b stock condition (A) and in the 96 FORMOscreen® conditions (B). Traces are colored based on the melting temperatures shown in Fig. 3.
Pre-formulation screening of a non-Ig protein
HIV-1 Env trimer
The trimeric HIV-1 envelope glycoprotein (Env) is the sole antigen presented to the human immune system during early HIV infection (Sanders et al. 2013). Antibodies binding to it (such as PGT145) can prevent infection by virus neutralization. A key feature of a HIV vaccine would hence be a stabilized Env. Such an Env-based HIV vaccine candidate, currently moving into clinical trials, is the BG505-SOSIP trimer.
Thermal stability
The BG505-SOSIP trimer displays a clean, single unfolding transition in its stock condition with a melting temperature of 65.7 °C (Fig. 5A), which is consistent with its described thermostability of 68.1 °C (Sanders et al. 2013). The FORMOscreen® conditions result in melting temperatures between 55.2 and 72.5 °C (Fig. 5B). Thus, the Env trimer shows a strong response of 17.3 °C to changes in buffer composition.
The screening suggests the optimal buffer for thermal stability is a citrate buffer at pH 6.5 with ~0.4 mg/mL polysorbate-80, and ~80 mg/mL sucrose. In comparison to the stock buffer, it results in a shift of + 6.8 °C. Buffers with low pH should be avoided in order to not reduce the thermal stability by up to ‑ 10.5 °C. This analysis highlights the potential of the FORMOscreen® for concurrently stabilizing antibodies and their cognate antigens.
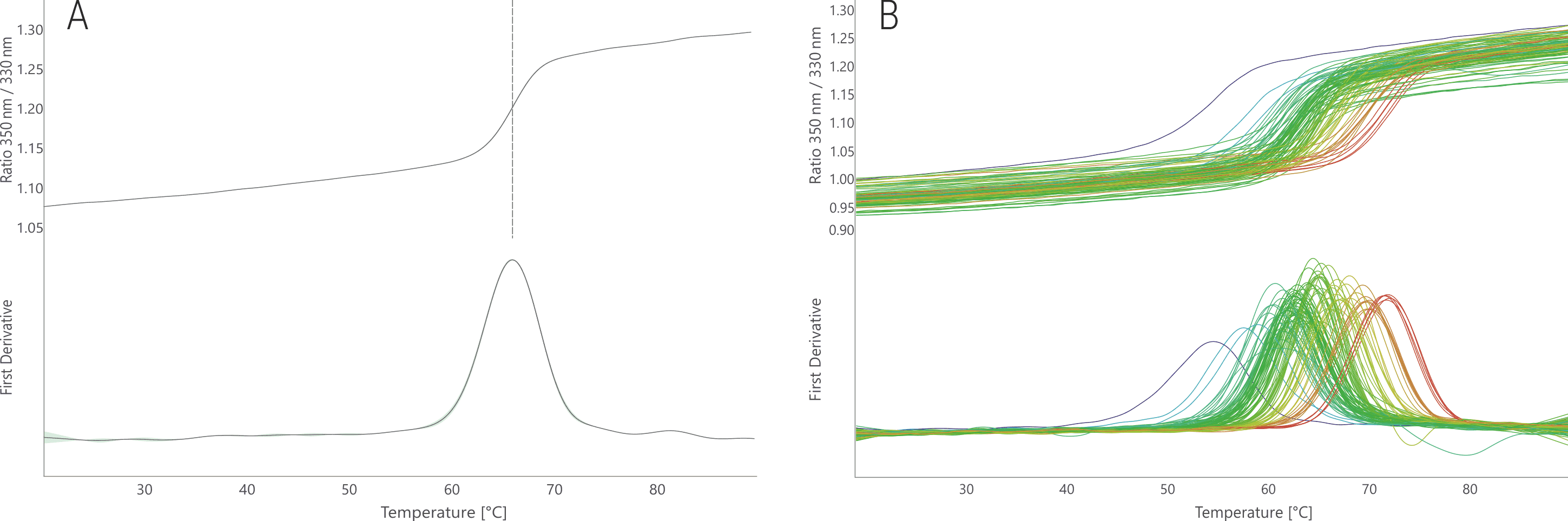
Figure 5. NanoDSF thermal melting of BG505-SOSIP stock condition (A) and in the 96 FORMOscreen® conditions (B). The upper panels show the raw fluorescence signal (350/330 nm ratio); the lower panels show the first derivative curves. Unfolding traces are colored from low (blue) to high (red) melting temperatures.
Aggregation
In its stock condition, no clear aggregation signal was observed for BG505-SOSIP (Fig. 6A). This can be due to the formation of very small and soluble aggregates or because the aggregate concentration was below the detection limit of the nanoDSF backscattering array. Nonetheless, many tested conditions resulted in a defined aggregation signal between 71 and 75 °C (Fig. 6B). As observed for the PGT145 antibody, also for BG505-SOSIP buffers leading to high thermal stability resulted in lower thermal aggregation onset temperatures.
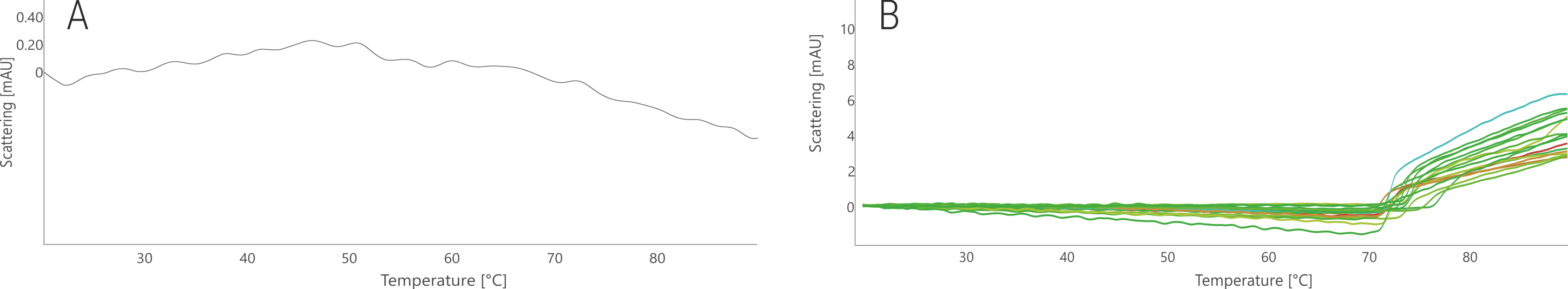
Figure 6. NanoDSF thermal aggregation analysis of BG505-SOSIP stock condition (A) and in the 96 FORMOscreen® conditions (B). Traces are colored based on the melting temperatures shown in Fig. 5.
2bind Services
2bind offers a nanoDSF-based stability and pre-formulation screening service using the FORMOscreen®, including detailed data analysis and ranking of buffer conditions. This service allows for rapid characterization of any antibody or protein of choice with respect to folding state, thermal stability and unfolding, as well as protein aggregation. The 2bind nanoDSF service guarantees minimal sample consumption requiring typically only 300 µL of a 1 mg/mL antibody solution for the analysis of all 96 FORMOscreen® buffers in duplicate measurements. 2bind is the world’s first service provider certified by NanoTemper Technologies for nanoDSF.
Additionally, 2bind offers MicroScale Thermophoresis (MST) based characterization of target-ligand interactions in the 96 FORMOscreen® conditions (steady-state affinity), as well as kinetic and thermodynamic analysis using Biolayer Interferometry (BLI) and Isothermal Titration Calorimetry (ITC).
nanoDSF Technology
NanoDSF is a variant of differential scanning fluorimetry for determining protein stability, unfolding, and aggregation under different thermal and chemical conditions. The truly label-free technique works in small glass capillaries, ensuring minimal sample consumption (Fig. 7A). NanoDSF uses protein-intrinsic tryptophan and tyrosine fluorescence, which changes upon thermal unfolding (Fig. 7B). From thermally or chemically induced shifts in the 330 nm and/or 350 nm fluorescence of a protein, the thermal melting point (Tm) or the chemical stability can be inferred (Fig. 7C). Importantly, samples can be studied without the use of a dye and with free choice of buffers. Melting temperatures of proteins with a concentration between 5 µg/ml and 250 mg/ml can be analyzed. In order to obtain high quality aggregation onset temperatures, protein solutions with concentrations above 1 mg/ml are required.
Since protein aggregation is a considerable point in pre- and final formulation, the special Prometheus NT.48 nanoDSF device available at 2bind features also back-reflection optics for aggregate detection. Normally, visible light passes through the capillaries containing the protein sample of interest without any interference. The light is reflected by a mirror on the capillary tray and gets finally quantified by the detector. The loss of reflection intensity is a precise measure for protein aggregation, which gets monitored in parallel to the intrinsic fluorescence.
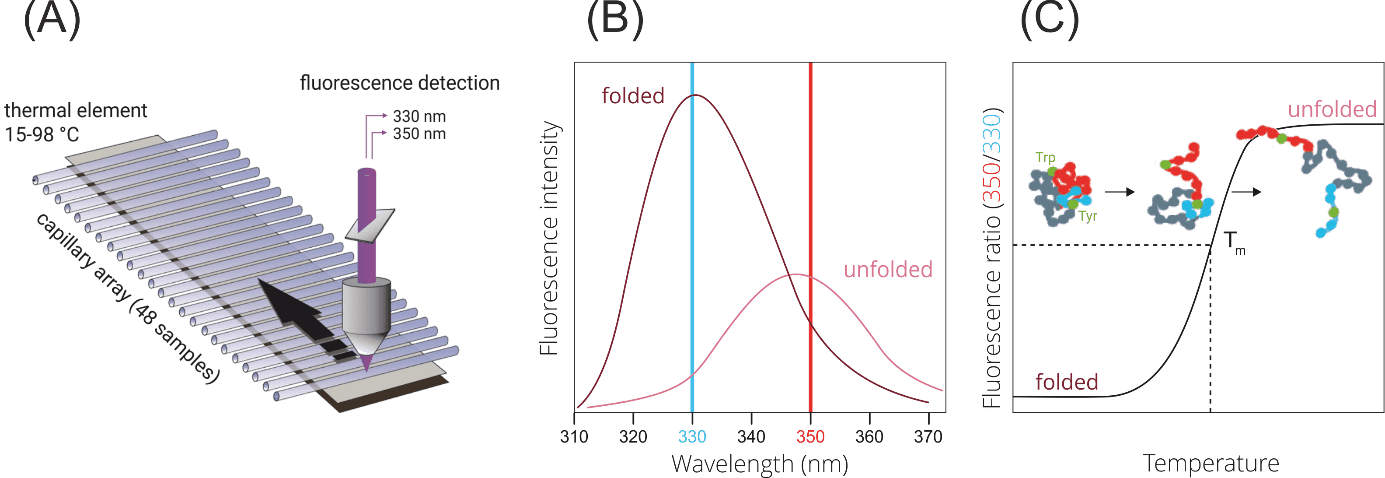
Figure 7. NanoDSF. (A) Technical setup. A fluorescence detection optic scans up to 48 capillaries in parallel, which are heated by a thermal element.(B) Fluorescence shifts upon protein denaturation. (C) Determination of the thermal melting temperature.
Literature
Grand view research. Biopharmaceuticals Contract Manufacturing Market Analysis, 2018 – 2025. GVR-2-68038-698-1 (2018).
Kwong et al. Structure of an HIV gp 120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998).
Lee et al. A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic β-Hairpin Structure. Immunity 46, 690–702 (2017).
Sanders et al. A Next-Generation Cleaved, Soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, Expresses Multiple Epitopes for Broadly Neutralizing but Not Non-Neutralizing Antibodies. PLoS Pathog 9(9): e1003618. doi:10.1371/journal.ppat.1003618 (2013).
Statista. Projected annual global biologics revenue from 2016 to 2022 (in billion U.S. dollars). https://www.statista.com/statistics/817593/revenue-forecast-for-global-biologics-market/. Accessed 2019-10-18.
Thali et al. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. Journal of Virology. 67, 3978–88 (1993).
Walker et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470 (2011).
Walsh. Biopharmaceutical benchmarks 2018. Nature Biotechnology 36, 1136-1145 (2018)
