Binding Kinetics of a Virus-like Particle Ebola Glycoprotein
and an Antibody Fab-fragment measured with Biolayer Interferometry (BLI)
Benjamin Zimmer, Corinna Popp, Maximilian Plach
2bind GmbH, Regensburg, Germany
BLI Technology
Biolayer Interferometry (BLI) measures binding kinetics. To do this, white light is sent down an optical glass fiber and is reflected from two optical layers (Figure 1, left): First, an internal reference layer, where reflection occurs at the interface between the glass fiber and the optical layer. Second, a bio-compatible layer, where reflection occurs at the interface with the surrounding liquid. Reflected light interferes either constructively or destructively at different wavelengths, resulting in a distinctive interference. While the light from the reference layer remains constant throughout an experiment, the attachment or detachment of molecules to/from the sensor surface changes the thickness of the bio-compatible layer and thereby the path length of the reflected light, resulting a spectral shift in the interference pattern (Figure 1, right) (Concepcion et al., 2009; Fortebio Application Note 14).
Therefore, BLI allows for observing target-ligand association and dissociation in real-time. Information from the binding curves can be used to determine binding parameters such as the association and dissociation rates constants (ka, kd) or the equilibrium dissociation constant (KD).
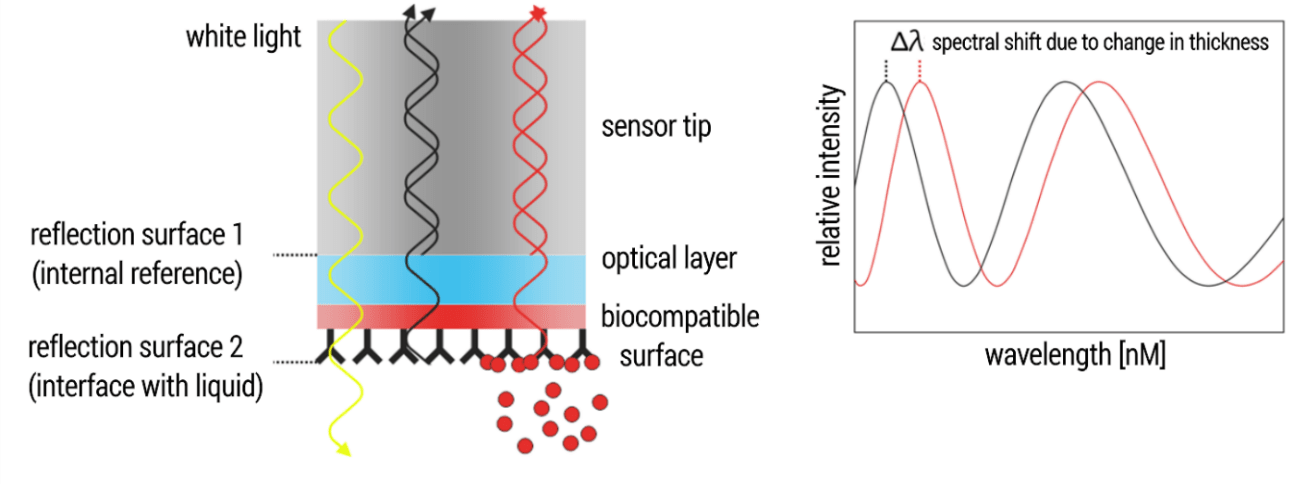
Figure 1. Technical BLI setup.
Introduction: Binding kinetics of virus surface proteins
A crucial step in the development of therapeutic antibodies is the assessment of their binding characteristics towards the desired antigen target. Ideally, this analysis takes place under most realistic conditions, i.e. not with simple, soluble antigens but with a more native-like form. Here, we demonstrate how BLI can be used to investigate the binding of an antibody Fab-fragment to an Ebola surface protein. However, not to a soluble variant but to the surface of an Ebola virus-like particle (EBOV VLP).
Ebola glycoprotein interactions
The passive administration of antibodies has great potential for treatment of severe viral diseases such as Ebola. Several studies demonstrated that monoclonal antibodies (mAbs) targeting the Ebola glycoprotein (GP) represent one of the most promising approaches for the treatment of this otherwise often lethal disease (Davidson et al.; 2015). The precise characterization of potential therapeutic antibodies always includes information about binding kinetics and affinities. Ideally, these parameters are determined under close-to-native conditions in order to obtain most realistic data. In case of Ebola GP this means to determine the binding kinetics of an antibody candidate towards the native, virus-particle expressed Ebola GP. Using a model system consisting of EBOV VLPs expressing Ebola GP and a Fab fragment of a humanized anti-Ebola GP antibody (h-13F6, Wilson et al. 2000), we show that BLI is a suitable technique for characterizing the kinetics and affinities of antibodies towards viral surface receptors in their membrane-bound, virus-particle presented state.
Assay set up
EBOV VLPs comprise high numbers of the highly glycosylated Ebola GP. For this reason, carbohydrate-binding proteins such as lectins can be used to capture EBOV-VLPs to the surface of BLI biosensors. In this study, initially biotinylated Wheat Germ Agglutinin (WGA) was immobilized to the surface of Streptavidin biosensors. This setup was then used for capturing the EBOV-VLPs (Figure 2). Notably, this strategy enabled a very stable capture of EBOV VLPs since the high numbers of GPs per VLP favors binding of the VLP to multiple WGA molecules, strengthening binding by avidity effects. After baseline establishment, the association and dissociation kinetics of Fab h-13F6 were analyzed (Figure 2).
Figure 2. Experimental setup for kinetic characterization of the interaction between EBOV VLP and Fab 13F6.
Results: Ebola VLP binding kinetics
For kinetic characterization, Fab h-13F6 was titrated to captured EBOV VLPs in a serial 1:3 dilution with a starting concentration of 300 nM (Figure 3A). Data were processed by reference well subtraction and a 1:1 interaction model was applied to obtain the binding affinity as well as the kinetic parameters of the interaction (Figure 3C). Overall, high quality data were obtained, indicated (i) the single-exponential association phase of the sensorgram traces which reflect the expected 1:1 binding situation between Ebola GP and the Fab and thus fit well to the applied 1:1 interaction model, (ii) low residuals in the range of the instrument noise (0.01 nm; Figure 3B), and (iii) good fit parameters (low X2-values and R2 > 0.95; Figure 3C).
Figure 3. Interaction between EBOV-VLP presented Ebola GP and Fab 13F6. (A) Sensorgram traces of a representative experiment (multi-cycle kinetic) with fit curves in red. (B) Residual plot. (C) Kinetic parameters obtained by fitting processed data with the 1:1 binding kinetics model (global fit). Values represent the mean ± standard deviation of two independent experiments.
Conclusion: 2bind BLI assay for measuring binding kinetics of virus surface proteins
In this application note, we described the successful establishment of a BLI setup for the straightforward characterization of the binding kinetics of the VLP-presented Ebola GP to the antibody Fab h-13F6. Such an assay format can be applied for analyzing interactions between other enveloped viruses or VLPs and their interaction partners, given that they carry enough glycosylated viral- or host-cell proteins. Thus, our BLI setup is interesting for everyone working with virus-associated surface proteins. Click here for a full list of services for antibody analysis. If you like to directly contact us for any kind of service, use the contact form below.
Literature
Concepcion et al. Label-free detection of bio-molecular interactions using BioLayer interfero-metry for kinetic characterization. Comb. Chem. High Throughput Screen. 2009, 12:791–800.
Davidson et al. Mechanism of Binding to Ebola Virus Glycoprotein by the ZMapp, ZMAb, and MB-003 Cocktail Antibodies. J Virol. 2015, 89(21): 10982–10992.
Fortébio. Application Note 14: Biomolecular Binding Kinetics Assays on the Octet Platform. Available at: https://www.fortebio.com/. (Accessed: 8th March 2019)
Wilson et al. Epitopes involved in antibody-mediated protection from Ebola virus. Science 2000, 289(5458):1664-1666.
