heliX Biosensor
Unrivaled information content and flexibility for demanding applications ApplicationsTechnologyheliX: Versatile Tool for Demanding Binding Analysis

The heliX® biosensor, the ultimate tool for multi-parameter comprehensive biophysical analysis of molecular interactions and dynamics.
Measure anything from binding affinity, avidity, and dose-response data, to demanding multi-specific interactions and ternary complex analyses (e.g. PROTACs), all the way to conformational changes and enzymatic activity with one platform. Confidently analyze binding of any analyte of your choice to DNA, RNA, aptamers, proteins, peptides, compounds, viral particles or liposomes.
2bind covers all necessary sample preparation steps including efficient and precise DNA-protein conjugation with the proFIRE® system. Read more below on the heliX® biosensor!
Affinity
LIGAND BINDING
KD
Equilibrium dissociation constant. Can be obtained by kinetic or classical equilibrium binding analysis. Provides information about the strength but not the dynamics of an interaction.
Avidity
Functional affinity, representing the overall strength of an interaction. Influenced by the binding affinity, binding valency, and the structural arrangement of target and ligand.
EC50/IC50
Dose-response data. Ligand concentration that gives half-maximal response or half-maximal activity.
Kinetics
BINDING KINETICS
ka (kon)
Association rate constant. Provides information on how fast complexes form; can be used for KD determination.
kd (koff)
Dissociation rate constant. Provides information on how fast complexes dissociate; can be used for KD determination.
KD
Equilibrium dissociation constant. Can be obtained by kinetic or classical equilibrium binding analysis. Provides information about the strength but not the dynamics of an interaction.
Avidity
Functional affinity, representing the overall strength of an interaction. Influenced by the binding affinity, binding valency, and the structural arrangement of target and ligand.
Other
MULTI-SPECIFIC INTERACTIONS
Two-color detection
Single-step binding analysis for complex interactions. Multi-specific antibodies or specialized FRET-based nanostructures for ternary complexes (e.g. PROTACs, molecular glues).
CONFORMATION AND STRUCTURE
Relative sizing experiments
Relative changes in hydrodynamic radius can be detected and protein size (in kDa) can be estimated.
Conformational change
Measurement of %-change of molecular friction, folding/unfolding, or agglomeration.
THERMODYNAMICS
ΔH, ΔG, ΔS
Measurement of binding enthalpy ΔH and entropy of binding ΔS, as well as derived from that free enthalpy of binding ΔG.
ENZYMATIC ACTIVITY
kcat, KM, kcat/KM
On-line measurement of enzyme activity parameters, e.g. for polymerases or helicases.
The heliX Biosensor has an Unrivaled Application Range
The heliX® biosensor is a chip-based instrument that utilizes the switchSENSE® technology. The easy and fast customization of the biochips allows for the investigation of any target molecule, for example viral particles, proteins, peptides, nucleic acids, small molecules and fragments. The two measurement spots per chip facilitate multiplexing as well as real-time referencing.
switchSENSE® offers multiple measurement modes and has a unique application range expanding far beyond 1-on-1 kinetics.
Applications by molecular parameters
Affinity, kinetics
High resolution real-time kinetics covering a broad range of ka and kd. Applicable from weak to tight binders.
ka = 103 to >108 M-1s-1
kd = 10-6 to 30 s-1
KD = 50 fM – 1 mM
Avidity and cooperative binding
Measure and quantify avidity effects separately from monovalent affinities. Special features for bispecific molecules.
Ternary complex formation
Simultaneous binding of bispecific reagents to both targets can be observed in real time, taking advantage of an inducible FRET effect.
Size and conformation
A protein’s relative size can be determined and conformational changes can be detected in relative studies.
Polymerase kinetics
Enzyme kinetics of DNA/RNA polymerization can be studied in real time.
Applications by molecule type
Bi-specific antibodies
Bi-specific antibodies can be characterized in-depth in an easy and automated workflow by functionalizing the chip surface with the two different antibody target molecules. The cell surface can be mimicked by specifically adjusting the density of the two antigens on the chip and the two-color fluorescence-based readout signal allows for the differentiation between binary affinities and ternary avidity. Thus, binding mechanisms can be elucidated and potent therapeutic antibody formats can be developed.
PROTACs and bi-specific molecules
For even more complex bispecific binders influencing protein-protein-interactions, specialized Y-shaped DNA nanostructures can be used. Each arm of the Y-structure can be functionalized with a different target protein. The arms are color-coded with a FRET pair. Upon simultaneous binding of the bispecific molecule to both targets the Y-structure closes and brings the dyes into a closer, FRET sensitive, distance. The formation of ternary complexes can thereby be followed in real time, enabling the screening of PROTACs, molecular glues, and other bispecific molecules.
Small molecules
Conformation and relative sizing
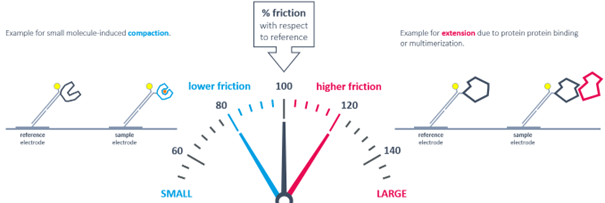
In the dynamic measurement mode, an alternating voltage is applied to the sensor surface, causing the DNA nanolevers to switch between upright and horizontal positions. The switching speed is influenced by the hydrodynamic diameter of the attached molecules. Thus, induced conformational changes like compaction or expansion can be detected and the size of the molecule can be estimated, offering an additional screening possibility for small molecules, ions, and more.
DNA, RNA, Polymerases
heliX® biosensor chips offer a unique opportunity to immobilize target DNA/RNA by hybridization with DNA anchors on the chip surface. Capturing of target DNA/RNA is possible by adding a single stranded overhang that hybridizes with the adapter sequences that anchor on the chip.
The setup for measuring hybridization of single stranded nucleic acids or kinetics of DNA/RNA binding molecules is therefore very simple and straight forward. Fluorescence proximity sensing enables the measurement of movements of enzymes on the DNA/RNA strand, and FRET has been successfully applied to monitor RNA tertiary structure formation.
Gene editing and manipulation of transcription and translation processes are highly relevant in science and therapy and biophysical characterization in heliX® biosensors delivers robust data in very fast measurements.
Potential applications include DNA-binding kinetics, CRISPR specificity, enzymatic activity and inhibitor screens, studies of DNA damage recognition and repair mechanisms, as well as RNA processing and regulation.
heliX Biosensor Measurement Modes
United in one device and using the switchSENSE® technology, Dynamic Biosensor’s heliX® biosensor delivers multi-parameter comprehensive biophysical analysis of molecular interactions and dynamics.
switchSENSE
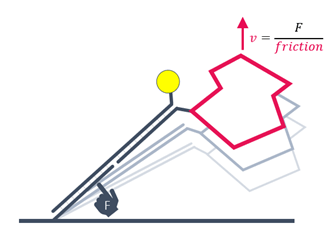
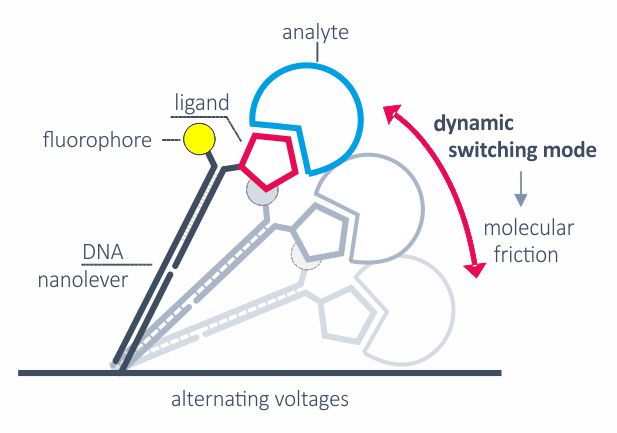
Alternating positive and negative voltage is applied to switch the DNA nanolevers from parallel position and proximity to the surface to an upright position. The switching speed is limited by hydrodynamic friction, which is defined by the size and shape of biomolecules. The switching mode can be applied to detect relative structural or conformational changes, for example from changes in assay conditions or from analyte binding.
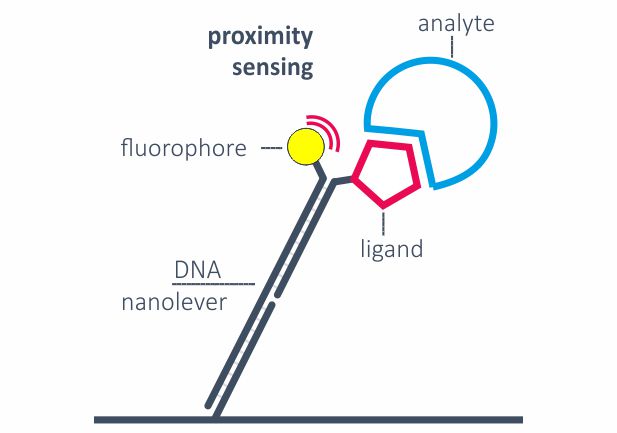
Fluorescence proximity sensing (FPS)
Real-time kinetics of binding events are typically monitored by Fluorescence proximity sensing (FPS). A constant negative voltage ensures that the DNA nanolevers are in an upright position and attached ligands keep sufficient distance from the surface to minimize mass-transport limitations. Two measurement spots, each with double-color detection, enable the parallel analysis of analytes binding to up to four ligands in parallel. FPS can also be applied to monitor molecular movements, for example the movement of a polymerase along the DNA.
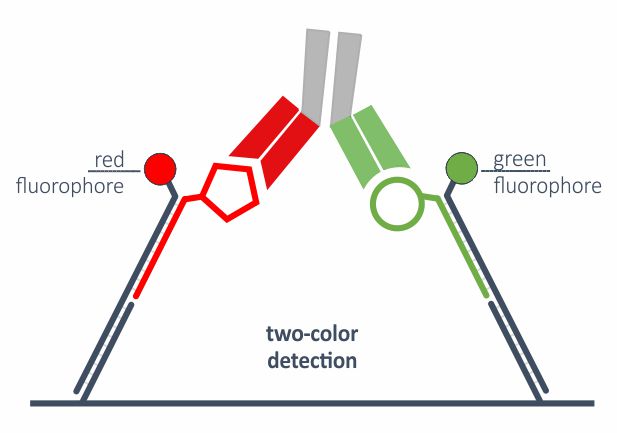
Two-color readout or FRET
Approximation of two molecules during dimerization or ternary complex formation can be studied via real-time FRET. Thich is enabled by advanced DNA nanostructures on the biosensor, two fluorophores and dual-color detection system. This allows for direct, one-step binding analysis of PROTACs or bi-specific antibodies, for example.
heliX Advantages
Broadest kinetic range
Quantify binding affinities (KD) from low pM to high µM with confident kinetic analysis.
High throughput
Temperature-controlled autosampler for 96- and 384-well plates. Automatic chip loader.
Advanced microfluidics
Disposable chips with integrated microfluidics and low maintenance guarantee precise and robust real-time kinetic measurements.
Measurement versatility
Three measurement modes (FPS, switchSENSE®, dual-color) cover basically any question in binding analytics.
Modularity
Several heliX+ instruments can be coupled to very high-throughput assays.
Technology
Overview
The switchSENSE® adapter biochips consist of a gold microelectrode to which DNA nanolevers are tethered. Due to the rigid structure of DNA double helices and their negative charge, their orientation on the chip surface can be controlled by applying an electrical potential to the electrode. Depending on the electrical potential, two different measurement modes can be differentiated. In the dynamic measurement mode, an alternating potential is used to cause an oscillating movement of the nanolevers. In the static measurement mode, a constant negative potential repels the nanolevers and keeps them in an upright orientation. Fluorescent dyes are attached at the distal ends of the DNA nanolevers. Single photon fluorescence counters allow for highly sensitive fluorescence detection of two different colours in parallel as read-out signals. Furthermore, the electrode contains two measurement spots, allowing for multiplexing of up to four different signals or real-time referencing. The desired ligand molecule will also be positioned at the end of the nanolever, whereas the analyte molecule will be flushed across the sensor surface in solution. In static measurement mode, a quenching of the fluorochrome is induced due to changes in the microenvironment of the dye. Once the analyte binds the ligand, collisional quenching and other processes cause a decrease in fluorescent signal intensity. This is also called fluorescence proximity sensing. In the dynamic measurement mode, the fluorochrome is quenched due to energy transfer processes to the surface whenever the dye is in close proximity to the gold electrode. The changes in fluorescence intensity are therefore directly linked to the switching speed of the nanolevers.
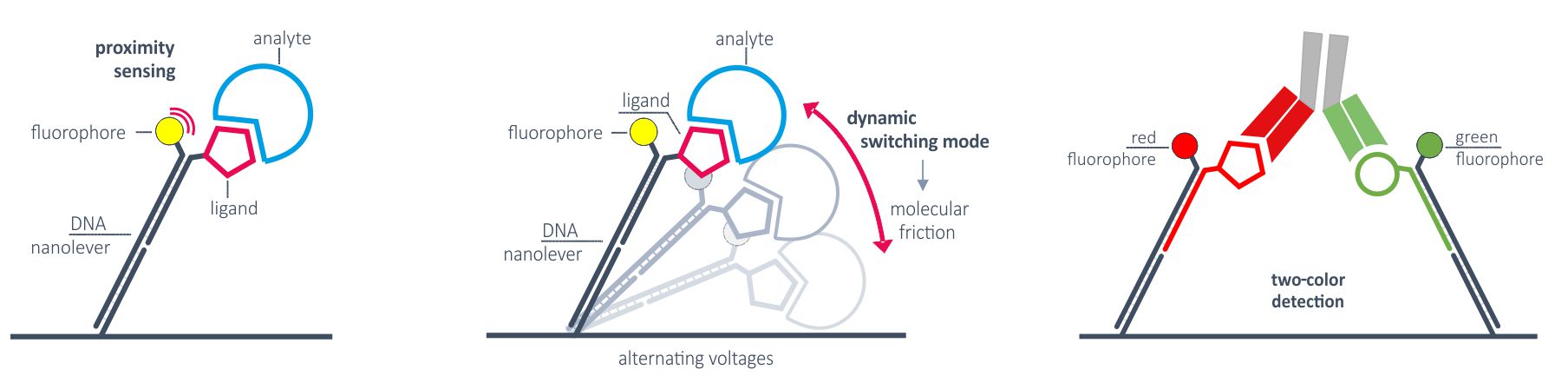
Applications of measurement modes
The static measurement mode/fluorescence proximity sensing can be used for high-performance kinetics of any molecular interaction. The broad detection range enables the investigation of very weak as well as very tight binders. The two colors allow for the differentiation of monovalent affinities and multivalent avidity to investigate for example bispecific antibodies.
Another unique feature of the static measurement mode is its ability to depict the catalytic activity of nucleic acid-modifying enzymes. It is possible to measure the activity of polymerases, transcriptases, helicases, etc in real-time and to separate their activity from their binding affinity.
The dynamic measurement mode can be used to detect conformational changes because these will change the hydrodynamic friction of the attached molecule, which affects the switching speed.
A third option how fluorescence can be quenched is by Förster resonance energy transfer (FRET). By combining two different fluorochromes on one nanolever, FRET can be induced and changes in FRET intensity can be related to binding events in more complex set-ups, e.g. for multi-domain proteins or to examine nucleic acid tertiary structure formation (e.g. aptamers). FRET technology is also the basis for investigations of proximity induced binding (e.g. PROTACs, molecular glues) with the Y-shaped DNA nanolevers specifically optimized for these applications.
Typical applications
The biochips offer endless opportunities for customized functionalizations
Kinetics applications
- Protein-protein interactions
- Receptor-ligand interactions
- Virus-like particle-peptide interactions
- Protein-small molecule interactions
- RNA/DNA-protein interactions
- RNA/DNA-small molecule interactions
- Transcription factors
- Splicing factors
Conformations applications
- Small molecules or ions inducing conformational changes
- Formation of multimeric protein complexes
- Posttranslational modifications
- Structure-function relationship
- Efficacy and mode of action of drugs
Multispecific Binders
- Multispecific antibodies binding mechanisms
- Optimization of antibody formats
- PROTACs
- Molecular glues
Nucleic acid targeting applications
- Screening of RNA targeting small molecules
- Polymerase/transcripase/helicase/etc inhibitor screening
- Small molecule – aptamer binding
- Riboswitches
- DNA damage repair
- Viral/bacterial transcription machinery
- CRISPR-Cas9
Frequently asked questions
FAQ – General
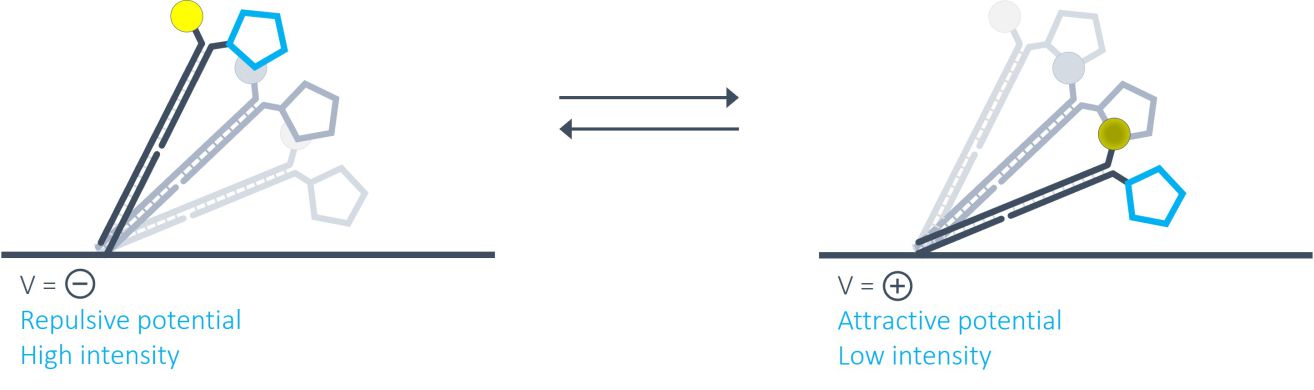
What is switchSENSE®? How does it work?
switchSENSE® is a novel biophysical research technology that works with electrically actuated DNA nanolevers on chip-based gold microelectrodes. In brief, by alternating electric potentials applied to the gold microelectrode one can either attract or repel the negatively charged backbones of double stranded DNA nanolevers grafted to the sensor surface. This generates an oscillating orientation change of the DNA nanolevers, called switching. By observing the fluorescence emission of a fluorescent dye at the distal end of the DNA nanolevers, it is possible to assess the position of the DNA nanolevers relative to the gold surface. The intensity of the fluorescent light emitted by the dye reports on the distance to the gold surface due to a distance dependent radiation-free energy transfer to the gold. In other words, the closer the fluorophore is positioned to the quenching gold surface, the less light it emits. Thus, at positive (attractive) potentials, the DNA nanolevers align horizontally with the electrode’s surface, positioning the fluorescent dye close to the gold surface resulting in low fluorescence intensity. On reversing the electrode potential to a negative (repulsive) potential, the DNA nanolevers gradually move into an upright orientation, which is accompanied by a gradual increase of fluorescence intensity. A schematic of the DNA switching process is shown in the figure below.
Why are heliX® biosensors particularly suited for measuring very fast and very slow kinetics?
The heliX® fluidic system has been designed as best-in-class to enable the quantitation of fast and slow kinetics (10+1 – 10-6 s-1) with confidence. Typically, kinetics are measured with nanolevers in an upright position (static mode). Binding events are detected by fluorescence proximity sensing.
Single-photon-counting detection for highest fluorescence sensitivity and data collection at 10 ms resolve even the fastest kinetics in real-time. High flow rates up to 500 µL/min eliminate mass-transport limitations and rebinding artefacts and thereby improve the measurement of rapid kinetics. Signals are stable and dissociations can be measured for up to 5000 min to quantify slowest dissociation rates.
Each of the two electrodes is equipped with a dual color detector and interactions of one analyte with four ligands can be measured in parallel.
Any constant temperature from 15°C to 40°C can be chosen for real time kinetic measurements.
How can the switchSENSE® technology be used to detect changes in conformation and size?
The measured instantaneous velocity of the nanolever depends on the hydrodynamic friction of the analyte.
Conformational changes are detectable by comparing to the initial state. Compaction results in reduced friction, whereas extension due to analyte binding or multimerization leads to increased friction.
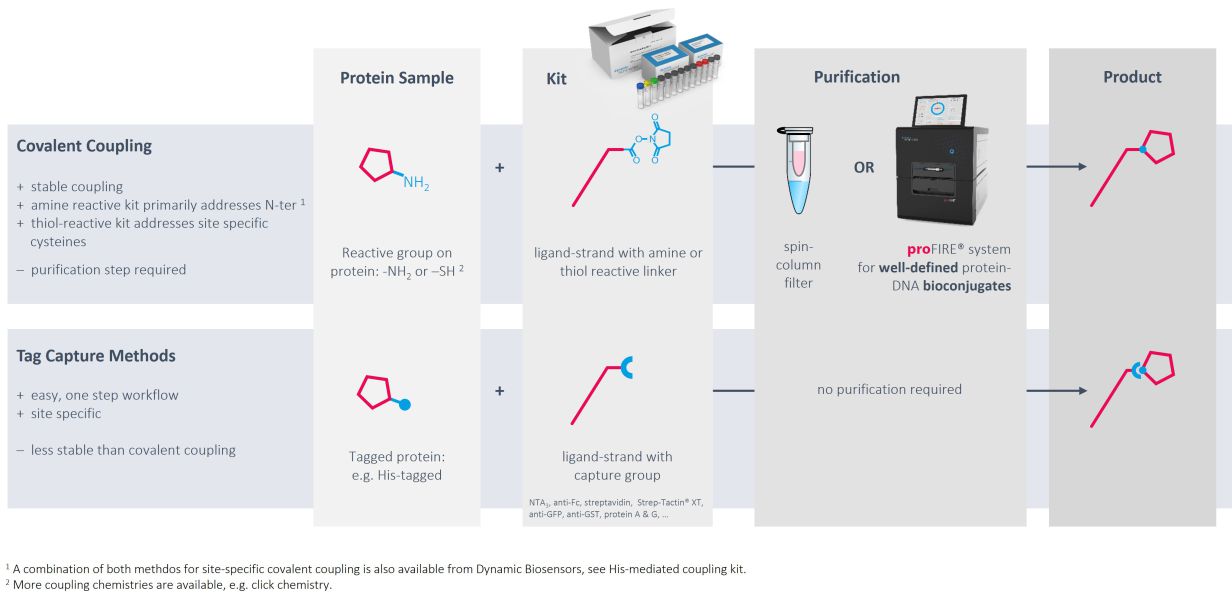
What options do I have to immobilize ligands on the heliX chip?
Key questions of assay design typically include the immobilization strategy. heliX® biosensor chips are equipped with single stranded anchor sequences and the most convenient way is the immobilization of target molecules via DNA hybridization. Different anchor sequences attached to electrode 1 and electrode 2 enable targeted immobilization to each spot. Typically, adapter strands carrying the fluorophore and ligand strands are prehybridized. Each electrode is equipped with a two-color detector and interactions of one analyte with up to four target molecules can be measured in parallel.
Immobilization via DNA hybridization is usually the method of choice to immobilize target DNA or RNA sequences. However, convenient, gentle and controlled conjugation methods as well as the proFIRE® purifier streamline the production of protein-DNA conjugates with high quality and purity, which can be easily implemented into the heliX® workflow.
Ready-to-use kits for coupling to primary amines exist for molecules with usual pIs as well as for low pI molecules. If the target of interest contains a His-tag, coupling can be directed to amines in proximity of the tag.
Alternatively, DNA strands can be coupled to thiol groups of free cysteines with high efficiencies and recovery rates.
As an alternative to chemical modifications of the targets of interest, tag-based capturing can be used. Methods and tools for capturing via His tags, biotin, twin-strep-tag, Fc parts and GFP have been developed and are ready to use.
What’s so cool about DNA-targeted immobilization?
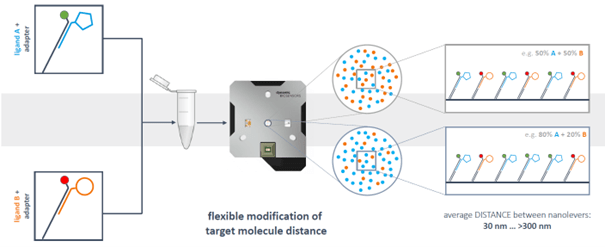
Actually, one chip type allows multiple assay designs. The chips are equipped with unique anchoring sequences in electrode 1 and electrode 2.
Dependent on the requirements of a specific experiment, the anchoring sequences can be hybridized with adapter strands of different shape in a functionalization step (see section “What types of adapter strands have been designed up to now?”). The DNA can be conjugated with the ligand, or with a capture molecule.
Via the ligand strand sequence, two different target molecules can be applied in the same functionalization step and directed to the two different electrodes. This is useful for measurement of a real-time reference.
However, DNA-targeting also enables full control of immobilization densities and spatial distribution of immobilized binding partners on each electrode. Anchoring DNA sequences are fixed on the electrodes in a defined distance. By diluting the fluorescently labeled ligand strand with ligand-free strand hybridized with adapter strand without fluorophore, the density of target on the surface can be reduced and the distance between target molecules can be increased in a highly controlled and reproducible manner.
Obviously, it is also possible to load two antigens on the surface. Prehybridization with the alternative fluorophore enables the measurement of interaction of an analyte with two target molecules on the surface simultaneously. The same is true for electrode 2, leading to a total number of four interactions that can be measured in parallel.
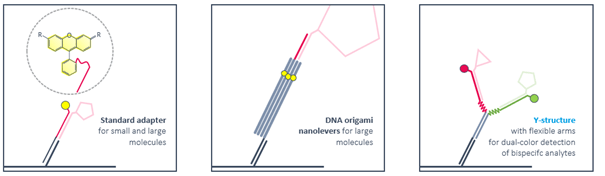
What types of adapter strands have been designed up to now?
Ligand strands are compatible with standard adapters as well as with DNA nanostructures. Rigid DNA-origami nanolevers are recommended for sizing experiments with large molecules. Antibody-like Y-structures with flexible arms reduce the distance between attached ligands and enable bivalent binding of smaller bispecific molecules like Protacs. Ternary complex formation and dimerization can be followed by real-time FRET.
What dyes can be used for proximity sensing?
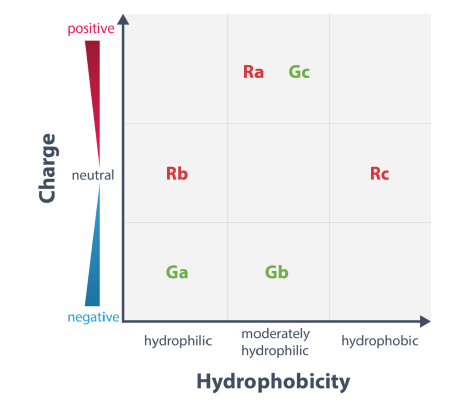
The two standard fluorescent probes for heliX® are the red and green dye Ra and Ga, respectively.
Fluorescence proximity sensing is based on the change in the local environment of the dye upon binding of an analyte to the ligand, which in turn results in a change of the fluorescence signal. This effect depends on the chemical nature of the dye and the interaction partner.
Therefore, a different dye may yield a higher signal response depending on the type of interaction. Dye scouting enables to screen for the most sensitive fluorophore for the respective application.
Three red and three green fluorophores with different chemical properties are available for dye scouting. The dyes differ in net charge and hydrophobicity as depicted below.
heliX® is equipped with LEDs and lasers to excite at 485 to 515 nm, or 600 to 630 nm, and to detect emissions at 520 to 580 nm, or 650 to 690 nm. Any dye respecting those characteristic can be used in heliX® applications.